Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica
- PMID: 17483225
- PMCID: PMC1913436
- DOI: 10.1128/JB.00451-07
Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica
Abstract
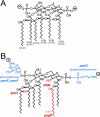
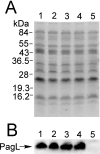
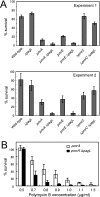
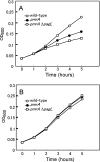
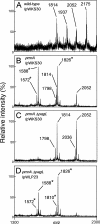
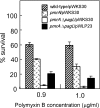
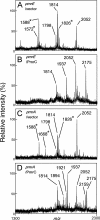
Salmonella enterica modifies its lipopolysaccharide (LPS), including the lipid A portion, to adapt to its environments. The lipid A 3-O-deacylase PagL exhibits latency; deacylation of lipid A is not usually observed in vivo despite the expression of PagL, which is under the control of a two-component regulatory system, PhoP-PhoQ. In contrast, PagL is released from latency in pmrA and pmrE mutants, both of which are deficient in aminoarabinose-modified lipid A, although the biological significance of this is not clear. The attachment of aminoarabinose to lipid A decreases the net anionic charge at the membrane's surface and reduces electrostatic repulsion between neighboring LPS molecules, leading to increases in bacterial resistance to cationic antimicrobial peptides, including polymyxin B. Here we examined the effects of the release of PagL from latency on resistance to polymyxin B. The pmrA pagL and pmrE pagL double mutants were more susceptible to polymyxin B than were the parental pmrA and pmrE mutants, respectively. Furthermore, introduction of the PagL expression plasmid into the pmrA pagL double mutant increased the resistance to polymyxin B. In addition, PagL-dependent deacylation of lipid A was observed in a mutant in which lipid A could not be modified with phosphoethanolamine, which partly contributes to the PmrA-dependent resistance to polymyxin B. These results, taken together, suggest that the release of PagL from latency compensates for the loss of resistance to polymyxin B that is due to a lack of other modifications to LPS.
Figures

Similar articles
-
Inhibition of Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification.J Bacteriol. 2005 Apr;187(7):2448-57. doi: 10.1128/JB.187.7.2448-2457.2005. J Bacteriol. 2005. PMID: 15774888 Free PMC article.
-
Extracellular loops of lipid A 3-O-deacylase PagL are involved in recognition of aminoarabinose-based membrane modifications in Salmonella enterica serovar typhimurium.J Bacteriol. 2008 Aug;190(16):5597-606. doi: 10.1128/JB.00587-08. Epub 2008 Jun 20. J Bacteriol. 2008. PMID: 18567660 Free PMC article.
-
pmrA(Con) confers pmrHFIJKL-dependent EGTA and polymyxin resistance on msbB Salmonella by decorating lipid A with phosphoethanolamine.J Bacteriol. 2007 Jul;189(14):5161-9. doi: 10.1128/JB.01969-06. Epub 2007 Apr 20. J Bacteriol. 2007. PMID: 17449614 Free PMC article.
-
Resistance to polymyxins: Mechanisms, frequency and treatment options.Drug Resist Updat. 2010 Aug-Oct;13(4-5):132-8. doi: 10.1016/j.drup.2010.05.002. Epub 2010 Jun 17. Drug Resist Updat. 2010. PMID: 20843473 Review.
-
[Outer membrane remodeling of Salmonella typhimurium and host innate immunity].Yakugaku Zasshi. 2006 Dec;126(12):1227-34. doi: 10.1248/yakushi.126.1227. Yakugaku Zasshi. 2006. PMID: 17139148 Review. Japanese.
Cited by
-
Latency of the lipid A deacylase PagL is involved in producing a robust permeation barrier in the outer membrane of Salmonella enterica.J Bacteriol. 2010 Nov;192(21):5837-40. doi: 10.1128/JB.00758-10. Epub 2010 Sep 10. J Bacteriol. 2010. PMID: 20833808 Free PMC article.
-
Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria.Front Microbiol. 2014 Nov 26;5:643. doi: 10.3389/fmicb.2014.00643. eCollection 2014. Front Microbiol. 2014. PMID: 25505462 Free PMC article. Review.
-
Phosphate groups of lipid A are essential for Salmonella enterica serovar Typhimurium virulence and affect innate and adaptive immunity.Infect Immun. 2012 Sep;80(9):3215-24. doi: 10.1128/IAI.00123-12. Epub 2012 Jul 2. Infect Immun. 2012. PMID: 22753374 Free PMC article.
-
Remodeling of Lipid A in Pseudomonas syringae pv. phaseolicola In Vitro.Int J Mol Sci. 2022 Feb 11;23(4):1996. doi: 10.3390/ijms23041996. Int J Mol Sci. 2022. PMID: 35216122 Free PMC article.
-
Identification of ColR binding consensus and prediction of regulon of ColRS two-component system.BMC Mol Biol. 2009 May 16;10:46. doi: 10.1186/1471-2199-10-46. BMC Mol Biol. 2009. PMID: 19445690 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

