The extracytoplasmic function-type sigma factor SigM of Corynebacterium glutamicum ATCC 13032 is involved in transcription of disulfide stress-related genes
- PMID: 17483229
- PMCID: PMC1913457
- DOI: 10.1128/JB.00382-07
The extracytoplasmic function-type sigma factor SigM of Corynebacterium glutamicum ATCC 13032 is involved in transcription of disulfide stress-related genes
Abstract
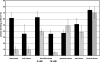
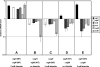
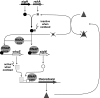
The gene for the extracytoplasmic function (ECF) sigma factor SigM was deleted from the chromosome of the gram-positive soil bacterium Corynebacterium glutamicum to elucidate the role of the SigM protein in the regulation of gene expression. Comparative DNA microarray hybridizations of the C. glutamicum wild type and sigM-deficient mutant C. glutamicum DN1 revealed 23 genes with enhanced expression in the sigM-proficient strain, encoding functions in the assembly of iron-sulfur clusters (suf operon), thioredoxin reductase (trxB), thioredoxins (trxC, trxB1), chaperones (groES, groEL, clpB), and proteins involved in the heat shock response (hspR, dnaJ, grpE). Deletion of the sigM gene rendered the C. glutamicum cells more sensitive to heat, cold, and the presence of the thiol oxidant diamide. Transcription of the sigM gene increased under different stress conditions, including heat shock, cold shock, and disulfide stress caused by diamide treatment, suggesting a regulatory role for SigM under thiol-oxidative stress conditions. Stress-responsive promoters were determined upstream of the suf operon and of the trxB, trxC, and trxB1 genes. The deduced SigM consensus promoter is characterized by the -35 hexamer gGGAAT and the -10 hexamer YGTTGR. Transcription of the sigM gene is apparently controlled by the ECF sigma factor SigH, since a sigH mutant was unable to enhance the expression of sigM and the SigM regulon under thiol-oxidative stress conditions. A typical SigH-responsive promoter was mapped upstream of the sigM gene. The ECF sigma factor SigM is apparently part of a regulatory cascade, and its transcription is controlled by SigH under conditions of thiol-oxidative stress.
Figures





Similar articles
-
Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network in Corynebacterium glutamicum.BMC Genomics. 2012 Sep 3;13:445. doi: 10.1186/1471-2164-13-445. BMC Genomics. 2012. PMID: 22943411 Free PMC article.
-
Functional analysis of sigH expression in Corynebacterium glutamicum.Biochem Biophys Res Commun. 2005 Jun 17;331(4):1542-7. doi: 10.1016/j.bbrc.2005.04.073. Biochem Biophys Res Commun. 2005. PMID: 15883048
-
The alternative sigma factor SigB of Corynebacterium glutamicum modulates global gene expression during transition from exponential growth to stationary phase.BMC Genomics. 2007 Jan 4;8:4. doi: 10.1186/1471-2164-8-4. BMC Genomics. 2007. PMID: 17204139 Free PMC article.
-
Sigma factors and promoters in Corynebacterium glutamicum.J Biotechnol. 2011 Jul 10;154(2-3):101-13. doi: 10.1016/j.jbiotec.2011.01.017. Epub 2011 Jan 26. J Biotechnol. 2011. PMID: 21277915 Review.
-
Analysis of Corynebacterium glutamicum promoters and their applications.Subcell Biochem. 2012;64:203-21. doi: 10.1007/978-94-007-5055-5_10. Subcell Biochem. 2012. PMID: 23080252 Review.
Cited by
-
Corynebacterium glutamicum promoters: a practical approach.Microb Biotechnol. 2013 Mar;6(2):103-17. doi: 10.1111/1751-7915.12019. Epub 2013 Jan 10. Microb Biotechnol. 2013. PMID: 23305350 Free PMC article. Review.
-
Exploring the role of sigma factor gene expression on production by Corynebacterium glutamicum: sigma factor H and FMN as example.Front Microbiol. 2015 Jul 22;6:740. doi: 10.3389/fmicb.2015.00740. eCollection 2015. Front Microbiol. 2015. PMID: 26257719 Free PMC article.
-
RosR (Cg1324), a hydrogen peroxide-sensitive MarR-type transcriptional regulator of Corynebacterium glutamicum.J Biol Chem. 2010 Sep 17;285(38):29305-18. doi: 10.1074/jbc.M110.156372. Epub 2010 Jul 19. J Biol Chem. 2010. PMID: 20643656 Free PMC article.
-
Induction of glutamic acid production by copper in Corynebacterium glutamicum.Appl Microbiol Biotechnol. 2021 Sep;105(18):6909-6920. doi: 10.1007/s00253-021-11516-3. Epub 2021 Aug 31. Appl Microbiol Biotechnol. 2021. PMID: 34463802
-
Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network in Corynebacterium glutamicum.BMC Genomics. 2012 Sep 3;13:445. doi: 10.1186/1471-2164-13-445. BMC Genomics. 2012. PMID: 22943411 Free PMC article.
References
-
- Aslund, F., and J. Beckwith. 1999. Bridge over troubled waters: sensing stress by disulfide bond formation. Cell 96:751-753. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

