Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment
- PMID: 17483276
- PMCID: PMC1932708
- DOI: 10.1128/AEM.02788-06
Tracking host sources of Cryptosporidium spp. in raw water for improved health risk assessment
Abstract
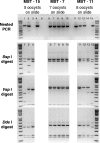
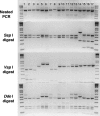
Recent molecular evidence suggests that different species and/or genotypes of Cryptosporidium display strong host specificity, altering our perceptions regarding the zoonotic potential of this parasite. Molecular forensic profiling of the small-subunit rRNA gene from oocysts enumerated on microscope slides by U.S. Environmental Protection Agency method 1623 was used to identify the range and prevalence of Cryptosporidium species and genotypes in the South Nation watershed in Ontario, Canada. Fourteen sites within the watershed were monitored weekly for 10 weeks to assess the occurrence, molecular composition, and host sources of Cryptosporidium parasites impacting water within the region. Cryptosporidium andersoni, Cryptosporidium muskrat genotype II, Cryptosporidium cervine genotype, C. baileyi, C. parvum, Cryptosporidium muskrat genotype I, the Cryptosporidium fox genotype, genotype W1, and genotype W12 were detected in the watershed. The molecular composition of the Cryptosporidium parasites, supported by general land use analysis, indicated that mature cattle were likely the main source of contamination of the watershed. Deer, muskrats, voles, birds, and other wildlife species, in addition to sewage (human or agricultural) may also potentially impact water quality within the study area. Source water protection studies that use land use analysis with molecular genotyping of Cryptosporidium parasites may provide a more robust source-tracking tool to characterize fecal impacts in a watershed. Moreover, the information is vital for assessing environmental and human health risks posed by water contaminated with zoonotic and/or anthroponotic forms of Cryptosporidium.
Figures



Similar articles
-
Cryptosporidium genotypes in wildlife from a new york watershed.Appl Environ Microbiol. 2007 Oct;73(20):6475-83. doi: 10.1128/AEM.01034-07. Epub 2007 Aug 24. Appl Environ Microbiol. 2007. PMID: 17720824 Free PMC article.
-
Molecular and phylogenetic approaches for assessing sources of Cryptosporidium contamination in water.Water Res. 2012 Oct 15;46(16):5135-50. doi: 10.1016/j.watres.2012.06.045. Epub 2012 Jul 10. Water Res. 2012. PMID: 22841595
-
Detection and differentiation of Cryptosporidium oocysts in water by PCR-RFLP.Methods Mol Biol. 2004;268:163-76. doi: 10.1385/1-59259-766-1:163. Methods Mol Biol. 2004. PMID: 15156028
-
Genotypes of Cryptosporidium from Sydney water catchment areas.J Appl Microbiol. 2005;98(5):1221-9. doi: 10.1111/j.1365-2672.2005.02562.x. J Appl Microbiol. 2005. PMID: 15836492
-
The European rabbit (Oryctolagus cuniculus), a source of zoonotic cryptosporidiosis.Zoonoses Public Health. 2010 Dec;57(7-8):e1-13. doi: 10.1111/j.1863-2378.2009.01308.x. Zoonoses Public Health. 2010. PMID: 20042061 Review.
Cited by
-
Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River watershed, Ontario, Canada.Appl Environ Microbiol. 2007 Sep;73(17):5401-10. doi: 10.1128/AEM.00354-07. Epub 2007 Jul 13. Appl Environ Microbiol. 2007. PMID: 17630309 Free PMC article.
-
Cryptosporidium genotypes in wildlife from a new york watershed.Appl Environ Microbiol. 2007 Oct;73(20):6475-83. doi: 10.1128/AEM.01034-07. Epub 2007 Aug 24. Appl Environ Microbiol. 2007. PMID: 17720824 Free PMC article.
-
Occurrences and genotypes of Cryptosporidium oocysts in river network of southern-eastern China.Parasitol Res. 2012 May;110(5):1701-9. doi: 10.1007/s00436-011-2688-6. Epub 2011 Oct 19. Parasitol Res. 2012. PMID: 22006191
-
Detection and resolution of Cryptosporidium species and species mixtures by genus-specific nested PCR-restriction fragment length polymorphism analysis, direct sequencing, and cloning.Appl Environ Microbiol. 2011 Jun;77(12):3998-4007. doi: 10.1128/AEM.02706-10. Epub 2011 Apr 15. Appl Environ Microbiol. 2011. PMID: 21498746 Free PMC article.
-
Occurrence, source, and human infection potential of cryptosporidium and Giardia spp. in source and tap water in shanghai, china.Appl Environ Microbiol. 2011 Jun;77(11):3609-16. doi: 10.1128/AEM.00146-11. Epub 2011 Apr 15. Appl Environ Microbiol. 2011. PMID: 21498768 Free PMC article.
References
-
- Boutros, S. N. 1989. Sampling and analysis for Cryptosporidium in PA public surface water supply sources. Pennsylvania Division of Water Supply, Harrisburg, PA.
-
- Brookes, J. D., M. R. Hipsey, M. D. Burch, R. H. Regel, L. G. Linden, C. M. Ferguson, and J. P. Antenucci. 2005. Relative value of surrogate indicators for detecting pathogens in lakes and reservoirs. Environ. Sci. Technol. 39:8614-8621. - PubMed
-
- Drinking Water Inspectorate. 2000. The water supply (water quality) regulations 2000. http://www.dwi.gov.uk/regs/si3184/3184.htm#273184. [Online.]
-
- Enemark, H. L., P. Ahrens, C. J. Lowery, S. M. Thamsborg, J. M. Enemark, V. Bille-Hansen, and P. Lind. 2002. Cryptosporidium andersoni from a Danish cattle herd: identification and preliminary characterisation. Vet. Parasitol. 107:37-49. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

