Specific detection and real-time PCR quantification of potentially mycophagous bacteria belonging to the genus Collimonas in different soil ecosystems
- PMID: 17483278
- PMCID: PMC1932782
- DOI: 10.1128/AEM.00387-07
Specific detection and real-time PCR quantification of potentially mycophagous bacteria belonging to the genus Collimonas in different soil ecosystems
Abstract
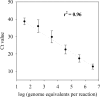
The bacterial genus Collimonas has the remarkable characteristic that it grows at the expense of living fungal hyphae under laboratory conditions. Here, we report the first field inventory of the occurrence and abundance of Collimonas in soils (n = 45) with naturally different fungal densities, which was performed in order to test the null hypothesis that there is a relationship between the presence of Collimonas and fungal biomass. Estimates of fungal densities were based on ergosterol measurements. Each soil was also characterized in terms of its physical and chemical properties and vegetation and management types. Culturable Collimonas was identified in plate-spread soil samples by its ability to clear colloidal chitin, in combination with a Collimonas-specific restriction fragment length polymorphism analysis of 16S rRNA PCR amplified from individual colonies. Using this approach, we found culturable collimonads only in (semi)natural grasslands. A real-time PCR assay for the specific quantification of Collimonas 16S rRNA in total soil DNA was developed. Collimonas was detectable in 80% of the soil samples, with densities up to 10(5) cells g(-1) (dry weight) soil. The numbers of Collimonas cells per gram of soil were consistently lowest in fungus-poor arable soils and, surprisingly, also in fungus-rich organic layers of forest soils. When all soils were included, no significant correlation was observed between the number of Collimonas cells and ergosterol-based soil fungal biomass. Based on this result, we rejected our null hypothesis, and possible explanations for this were addressed.
Figures



References
-
- Aspray, T. J., S. K. Hansen, and R. G. Burns. 2005. A soil-based microbial biofilm exposed to 2,4-D: bacterial community development and establishment of conjugative plasmid pJP4. FEMS Microbiol. Ecol. 54:317-327. - PubMed
-
- Bach, H. J., J. Tomanova, M. Schloter, and J. C. Munch. 2002. Enumeration of total bacteria and bacteria with genes for proteolytic activity in pure cultures and in environmental samples by quantitative PCR mediated amplification. J. Microbiol. Methods 49:235-245. - PubMed
-
- Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541-555. - PubMed
-
- Baker, R. 1987. Mycoparasitism—ecology and physiology. Can. J. Plant Pathol. 9:370-379.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

