Genetic dissection of parallel sister-chromatid cohesion pathways
- PMID: 17483413
- PMCID: PMC1931553
- DOI: 10.1534/genetics.107.072876
Genetic dissection of parallel sister-chromatid cohesion pathways
Abstract
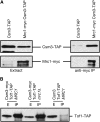
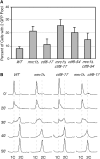
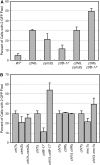
Sister-chromatid cohesion, the process of pairing replicated chromosomes during mitosis and meiosis, is mediated through the essential cohesin complex and a number of nonessential cohesion genes, but the specific roles of these nonessential genes in sister-chromatid cohesion remain to be clarified. We analyzed sister-chromatid cohesion in double mutants of mrc1Delta, tof1Delta, and csm3Delta and identified additive cohesion defects that indicated the existence of at least two pathways that contribute to sister-chromatid cohesion. To understand the relationship of other nonessential cohesion genes with respect to these two pathways, pairwise combinations of deletion and temperature-sensitive alleles were tested for cohesion defects. These data defined two cohesion pathways, one containing CSM3, TOF1, CTF4, and CHL1, and the second containing MRC1, CTF18, CTF8, and DCC1. Furthermore, we found that the nonessential genes are not important for the maintenance of cohesion at G(2)/M. Thus, our data suggest that nonessential cohesion genes make critical redundant contributions to the establishment of sister-chromatid cohesion and define two cohesion pathways, thereby establishing a framework for understanding the role of nonessential genes in sister-chromatid cohesion.
Figures






Similar articles
-
Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II.J Cell Sci. 2004 Jul 15;117(Pt 16):3547-59. doi: 10.1242/jcs.01231. Epub 2004 Jun 29. J Cell Sci. 2004. PMID: 15226378
-
An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae.Chromosoma. 2013 Mar;122(1-2):121-34. doi: 10.1007/s00412-013-0396-y. Epub 2013 Jan 20. Chromosoma. 2013. PMID: 23334284 Free PMC article.
-
Replisome-cohesin interactions provided by the Tof1-Csm3 and Mrc1 cohesion establishment factors.Chromosoma. 2023 Jun;132(2):117-135. doi: 10.1007/s00412-023-00797-4. Epub 2023 May 11. Chromosoma. 2023. PMID: 37166686 Free PMC article.
-
The Interplay of Cohesin and the Replisome at Processive and Stressed DNA Replication Forks.Cells. 2021 Dec 8;10(12):3455. doi: 10.3390/cells10123455. Cells. 2021. PMID: 34943967 Free PMC article. Review.
-
The molecular basis of sister-chromatid cohesion.Annu Rev Cell Dev Biol. 2001;17:753-77. doi: 10.1146/annurev.cellbio.17.1.753. Annu Rev Cell Dev Biol. 2001. PMID: 11687503 Review.
Cited by
-
Ctf18-RFC and DNA Pol ϵ form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication.Nucleic Acids Res. 2020 Aug 20;48(14):8128-8145. doi: 10.1093/nar/gkaa541. Nucleic Acids Res. 2020. PMID: 32585006 Free PMC article.
-
A genetic screen for replication initiation defective (rid) mutants in Schizosaccharomyces pombe.Cell Div. 2010 Aug 27;5:20. doi: 10.1186/1747-1028-5-20. Cell Div. 2010. PMID: 20799962 Free PMC article.
-
Histone H3 K56 hyperacetylation perturbs replisomes and causes DNA damage.Genetics. 2008 Aug;179(4):1769-84. doi: 10.1534/genetics.108.088914. Epub 2008 Jun 24. Genetics. 2008. PMID: 18579506 Free PMC article.
-
Chl1 helicase controls replication fork progression by regulating dNTP pools.Life Sci Alliance. 2022 Jan 11;5(4):e202101153. doi: 10.26508/lsa.202101153. Print 2022 Apr. Life Sci Alliance. 2022. PMID: 35017203 Free PMC article.
-
Chromosome segregation in budding yeast: sister chromatid cohesion and related mechanisms.Genetics. 2014 Jan;196(1):31-63. doi: 10.1534/genetics.112.145144. Genetics. 2014. PMID: 24395824 Free PMC article. Review.
References
-
- Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler et al., 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3: 958–965. - PubMed
-
- Baetz, K. K., N. J. Krogan, A. Emili, J. Greenblatt and P. Hieter, 2004. The ctf13–30/CTF13 genomic haploinsufficiency modifier screen identifies the yeast chromatin remodeling complex RSC, which is required for the establishment of sister chromatid cohesion. Mol. Cell. Biol. 24(3): 1232–1244. - PMC - PubMed
-
- Bailis, J. M., P. Bernard, R. Antonelli, R. C. Allshire and S. L. Forsburg, 2003. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 5: 1111–1116. - PubMed
-
- Barbour, L., and W. Xiao, 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 532: 137–155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

