Patterns of recombination and MLH1 foci density along mouse chromosomes: modeling effects of interference and obligate chiasma
- PMID: 17483430
- PMCID: PMC1931555
- DOI: 10.1534/genetics.106.070235
Patterns of recombination and MLH1 foci density along mouse chromosomes: modeling effects of interference and obligate chiasma
Abstract
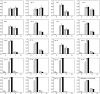
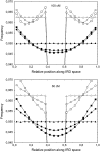
Crossover interference in meiosis is often modeled via stationary renewal processes. Here we consider a new model to incorporate the known biological feature of "obligate chiasma" whereby in most organisms each bivalent almost always has at least one crossover. The initial crossover is modeled as uniformly distributed along the chromosome, and starting from its position, subsequent crossovers are placed with forward and backward stationary renewal processes using a chi-square distribution of intercrossover distances. We used our model as well as the standard chi-square model to simulate the patterns of crossover densities along bivalents or chromatids for those having zero, one, two, or three or more crossovers; indeed, such patterns depend on the number of crossovers. With both models, simulated patterns compare very well to those found experimentally in mice, both for MLH1 foci on bivalents and for crossovers on genetic maps. However, our model provides a better fit to experimental data as compared to the standard chi-square model, particularly regarding the distribution of numbers of crossovers per chromosome. Finally, our model predicts an enhancement of the recombination rate near the extremities, which, however, explains only a part of the pattern observed in mouse.
Figures



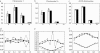

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

