Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing
- PMID: 17483454
- PMCID: PMC1876591
- DOI: 10.1073/pnas.0700865104
Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing
Abstract
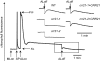
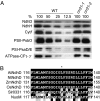
The pentatricopeptide repeat (PPR) proteins form one of the largest families in higher plants and are believed to be involved in the posttranscriptional processes of gene expression in plant organelles. It has been shown by using a genetic approach focusing on NAD(P)H dehydrogenase (NDH) activity that a PPR protein CRR4 is essential for a specific RNA editing event in chloroplasts. Here, we discovered Arabidopsis crr21 mutants that are specifically impaired in the RNA editing of the site 2 of ndhD (ndhD-2), which encodes a subunit of the NDH complex. The CRR21 gene encodes a member of the PPR protein family. The RNA editing of ndhD-2 converts the Ser-128 of NdhD to leucine. In crr21, the activity of the NDH complex is specifically impaired, suggesting that the Ser128Leu change has important consequences for the function of the NDH complex. Both CRR21 and CRR4 belong to the E+ subgroup in the PLS subfamily that is characterized by the presence of a conserved C-terminal region (the E/E+ domain). This E/E+ domain is highly conserved and exchangeable between CRR21 and CRR4, although it is not essential for the RNA binding. Our results suggest that the E/E+ domain has a common function in RNA editing rather than of recognizing specific RNA sequences.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
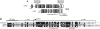

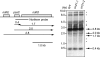

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

