Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes
- PMID: 17483479
- PMCID: PMC1890498
- DOI: 10.1073/pnas.0702741104
Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes
Abstract
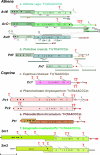
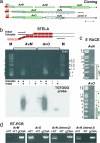
The evolutionary origin of telomerases, enzymes that maintain the ends of linear chromosomes in most eukaryotes, is a subject of debate. Penelope-like elements (PLEs) are a recently described class of eukaryotic retroelements characterized by a GIY-YIG endonuclease domain and by a reverse transcriptase domain with similarity to telomerases and group II introns. Here we report that a subset of PLEs found in bdelloid rotifers, basidiomycete fungi, stramenopiles, and plants, representing four different eukaryotic kingdoms, lack the endonuclease domain and are located at telomeres. The 5' truncated ends of these elements are telomere-oriented and typically capped by species-specific telomeric repeats. Most of them also carry several shorter stretches of telomeric repeats at or near their 3' ends, which could facilitate utilization of the telomeric G-rich 3' overhangs to prime reverse transcription. Many of these telomere-associated PLEs occupy a basal phylogenetic position close to the point of divergence from the telomerase-PLE common ancestor and may descend from the missing link between early eukaryotic retroelements and present-day telomerases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Comment in
-
The beginning of the end: links between ancient retroelements and modern telomerases.Proc Natl Acad Sci U S A. 2007 May 29;104(22):9107-8. doi: 10.1073/pnas.0703224104. Epub 2007 May 21. Proc Natl Acad Sci U S A. 2007. PMID: 17517612 Free PMC article. No abstract available.
References
-
- Blackburn EH. Nat Struct Biol. 2000;7:847–850. - PubMed
-
- Eickbush TH. Science. 1997;277:911–912. - PubMed
-
- Nakamura TM, Cech TR. Cell. 1998;92:587–590. - PubMed
-
- Arkhipova IR, Pyatkov KI, Meselson M, Evgen'ev MB. Nat Genet. 2003;33:123–124. - PubMed
-
- Kazazian HH., Jr Science. 2004;303:1626–1632. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

