Expression stability of commonly used reference genes in canine articular connective tissues
- PMID: 17484782
- PMCID: PMC1884148
- DOI: 10.1186/1746-6148-3-7
Expression stability of commonly used reference genes in canine articular connective tissues
Abstract
Background: The quantification of gene expression in tissue samples requires the use of reference genes to normalise transcript numbers between different samples. Reference gene stability may vary between different tissues, and between the same tissue in different disease states. We evaluated the stability of 9 reference genes commonly used in human gene expression studies. Real-time reverse transcription PCR and a mathematical algorithm were used to establish which reference genes were most stably expressed in normal and diseased canine articular tissues and two canine cell lines stimulated with lipolysaccaride (LPS).
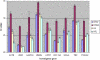
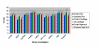
Results: The optimal reference genes for comparing gene expression data between normal and diseased infrapatella fat pad were RPL13A and YWHAZ (M = 0.56). The ideal reference genes for comparing normal and osteoarthritic (OA) cartilage were RPL13A and SDHA (M = 0.57). The best reference genes for comparing normal and ruptured canine cranial cruciate ligament were B2M and TBP (M = 0.59). The best reference genes for normalising gene expression data from normal and LPS stimulated cell lines were SDHA and YWHAZ (K6) or SDHA and HMBS (DH82), which had expression stability (M) values of 0.05 (K6) and 0.07 (DH82) respectively. The number of reference genes required to reduce pairwise variation (V) to <0.20 was 4 for cell lines, 5 for cartilage, 7 for cranial cruciate ligament and 8 for fat tissue. Reference gene stability was not related to the level of gene expression.
Conclusion: The reference genes demonstrating the most stable expression within each different canine articular tissue were identified, but no single reference gene was identified as having stable expression in all different tissue types. This study underlines the necessity to select reference genes on the basis of tissue and disease specific expression profile evaluation and highlights the requirement for the identification of new reference genes with greater expression stability for use in canine articular tissue gene expression studies.
Figures


References
-
- Aigner T, Zien A, Gehrsitz A, Gebhard PM, McKenna L. Anabolic and catabolic gene expression pattern analysis in normal versus osteoarthritic cartilage using complementary DNA-array technology. Arthritis Rheum. 2001;44:2777–2789. doi: 10.1002/1529-0131(200112)44:12<2777::AID-ART465>3.0.CO;2-H. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

