c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1
- PMID: 17485441
- PMCID: PMC1951955
- DOI: 10.1128/MCB.02282-06
c-Myc transforms human mammary epithelial cells through repression of the Wnt inhibitors DKK1 and SFRP1
Abstract
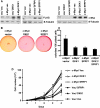
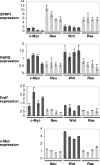
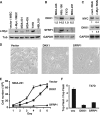
c-myc is frequently amplified in breast cancer; however, the mechanism of myc-induced mammary epithelial cell transformation has not been defined. We show that c-Myc induces a profound morphological transformation in human mammary epithelial cells and anchorage-independent growth. c-Myc suppresses the Wnt inhibitors DKK1 and SFRP1, and derepression of DKK1 or SFRP1 reduces Myc-dependent transforming activity. Myc-dependent repression of DKK1 and SFRP1 is accompanied by Wnt target gene activation and endogenous T-cell factor activity. Myc-induced mouse mammary tumors have repressed SFRP1 and increased expression of Wnt target genes. DKK1 and SFRP1 inhibit the transformed phenotype of breast cancer cell lines, and DKK1 inhibits tumor formation. We propose a positive feedback loop for activation of the c-myc and Wnt pathways in breast cancer.
Figures


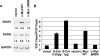

Similar articles
-
Turning the tables: Myc activates Wnt in breast cancer.Cell Cycle. 2007 Nov 1;6(21):2625-7. doi: 10.4161/cc.6.21.4880. Epub 2007 Aug 13. Cell Cycle. 2007. PMID: 17726380 Review.
-
Loss of SFRP1 promotes ductal branching in the murine mammary gland.BMC Dev Biol. 2012 Aug 28;12:25. doi: 10.1186/1471-213X-12-25. BMC Dev Biol. 2012. PMID: 22928951 Free PMC article.
-
WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth.Breast Cancer Res. 2009;11(3):R32. doi: 10.1186/bcr2317. Epub 2009 May 27. Breast Cancer Res. 2009. PMID: 19473496 Free PMC article.
-
Repressor roles for TCF-4 and Sfrp1 in Wnt signaling in breast cancer.Oncogene. 2006 Jul 20;25(31):4361-9. doi: 10.1038/sj.onc.1209470. Epub 2006 Mar 13. Oncogene. 2006. PMID: 16532032
-
Role of Secreted Frizzled-Related Protein 1 in Early Mammary Gland Tumorigenesis and Its Regulation in Breast Microenvironment.Cells. 2020 Jan 14;9(1):208. doi: 10.3390/cells9010208. Cells. 2020. PMID: 31947616 Free PMC article. Review.
Cited by
-
DNAJB6 governs a novel regulatory loop determining Wnt/β-catenin signalling activity.Biochem J. 2012 Jun 15;444(3):573-80. doi: 10.1042/BJ20120205. Biochem J. 2012. PMID: 22455953 Free PMC article.
-
Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis.Cell Mol Life Sci. 2019 Nov;76(21):4275-4289. doi: 10.1007/s00018-019-03222-1. Epub 2019 Jul 15. Cell Mol Life Sci. 2019. PMID: 31309249 Free PMC article. Review.
-
Pim-2 kinase is an important target of treatment for tumor progression and bone loss in myeloma.Leukemia. 2015 Jan;29(1):207-17. doi: 10.1038/leu.2014.147. Epub 2014 May 2. Leukemia. 2015. PMID: 24787487
-
The network of pluripotency, epithelial-mesenchymal transition, and prognosis of breast cancer.Breast Cancer (Dove Med Press). 2015 Sep 7;7:303-19. doi: 10.2147/BCTT.S71163. eCollection 2015. Breast Cancer (Dove Med Press). 2015. PMID: 26379447 Free PMC article. Review.
-
Combined detection of serum Dickkopf-1 and its autoantibodies to diagnose esophageal squamous cell carcinoma.Cancer Med. 2016 Jul;5(7):1388-96. doi: 10.1002/cam4.702. Epub 2016 Mar 14. Cancer Med. 2016. PMID: 26988995 Free PMC article.
References
-
- Bafico, A., A. Gazit, T. Pramila, P. W. Finch, A. Yaniv, and S. A. Aaronson. 1999. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J. Biol. Chem. 274:16180-16187. - PubMed
-
- Bafico, A., G. Liu, L. Goldin, V. Harris, and S. A. Aaronson. 2004. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell 6:497-506. - PubMed
-
- Baylin, S. B., and J. E. Ohm. 2006. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer 6:107-116. - PubMed
-
- Bejsovec, A. 2005. Wnt pathway activation: new relations and locations. Cell 120:11-14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
