The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation
- PMID: 17485514
- PMCID: PMC2118584
- DOI: 10.1084/jem.20062342
The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation
Abstract
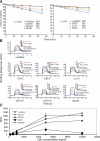
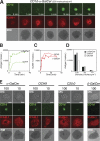
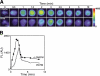
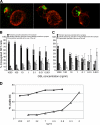
CD1d-restricted lymphocytes recognize a broad lipid range. However, how CD1d-restricted lymphocytes translate T cell receptor (TCR) recognition of lipids with similar group heads into distinct biological responses remains unclear. Using a soluble invariant NKT (iNKT) TCR and a newly engineered antibody specific for alpha-galactosylceramide (alpha-GalCer)-human CD1d (hCD1d) complexes, we measured the affinity of binding of iNKT TCR to hCD1d molecules loaded with a panel of alpha-GalCer analogues and assessed the rate of dissociation of alpha-GalCer and alpha-GalCer analogues from hCD1d molecules. We extended this analysis by studying iNKT cell synapse formation and iNKT cell activation by the same panel of alpha-GalCer analogues. Our results indicate the unique role of the lipid chain occupying the hCD1d F' channel in modulating TCR binding affinity to hCD1d-lipid complexes, the formation of stable immunological synapse, and cell activation. These data are consistent with previously described conformational changes between empty and loaded hCD1d molecules (Koch, M., V.S. Stronge, D. Shepherd, S.D. Gadola, B. Mathew, G. Ritter, A.R. Fersht, G.S. Besra, R.R. Schmidt, E.Y. Jones, and V. Cerundolo. 2005. Nat. Immunol 6:819-826), suggesting that incomplete occupation of the hCD1d F' channel results in conformational differences at the TCR recognition surface. This indirect effect provides a general mechanism by which lipid-specific lymphocytes are capable of recognizing both the group head and the length of lipid antigens, ensuring greater specificity of antigen recognition.
Figures


References
-
- Brigl, M., and M.B. Brenner. 2004. CD1: antigen presentation and T cell function. Annu. Rev. Immunol. 22:817–890. - PubMed
-
- Brutkiewicz, R.R. 2006. CD1d ligands: the good, the bad, and the ugly. J. Immunol. 177:769–775. - PubMed
-
- Kinjo, Y., E. Tupin, D. Wu, M. Fujio, R. Garcia-Navarro, M.R. Benhnia, D.M. Zajonc, G. Ben-Menachem, G.D. Ainge, G.F. Painter, et al. 2006. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat. Immunol. 7:978–986. - PubMed
-
- Koch, M., V.S. Stronge, D. Shepherd, S.D. Gadola, B. Mathew, G. Ritter, A.R. Fersht, G.S. Besra, R.R. Schmidt, E.Y. Jones, and V. Cerundolo. 2005. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat. Immunol. 6:819–826. - PubMed
-
- Zeng, Z., A.R. Castano, B.W. Segelke, E.A. Stura, P.A. Peterson, and I.A. Wilson. 1997. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 277:339–345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

