A semisynthetic epitope for kinase substrates
- PMID: 17486086
- PMCID: PMC2932705
- DOI: 10.1038/nmeth1048
A semisynthetic epitope for kinase substrates
Abstract
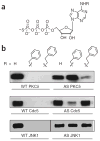
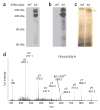
The ubiquitous nature of protein phosphorylation makes it challenging to map kinase-substrate relationships, which is a necessary step toward defining signaling network architecture. To trace the activity of individual kinases, we developed a semisynthetic reaction scheme, which results in the affinity tagging of substrates of the kinase in question. First, a kinase, engineered to use a bio-orthogonal ATPgammaS analog, catalyzes thiophosphorylation of its direct substrates. Second, alkylation of thiophosphorylated serine, threonine or tyrosine residues creates an epitope for thiophosphate ester-specific antibodies. We demonstrated the generality of semisynthetic epitope construction with 13 diverse kinases: JNK1, p38alpha MAPK, Erk1, Erk2, Akt1, PKCdelta, PKCepsilon, Cdk1/cyclinB, CK1, Cdc5, GSK3beta, Src and Abl. Application of this approach, in cells isolated from a mouse that expressed endogenous levels of an analog-specific (AS) kinase (Erk2), allowed purification of a direct Erk2 substrate.
Conflict of interest statement
Figures


References
-
- Berwick DC, Tavare JM. Identifying protein kinase substrates: hunting for the organ-grinder’s monkeys. Trends Biochem Sci. 2004;29:227–232. - PubMed
-
- Ptacek J, Snyder M. Charging it up: global analysis of protein phosphorylation. Trends Genet. 2006;22:545–554. - PubMed
-
- Ptacek J, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. - PubMed
-
- Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

