Molecular characterization of a mammalian smooth muscle myosin light chain kinase
- PMID: 1748666
- PMCID: PMC2836767
Molecular characterization of a mammalian smooth muscle myosin light chain kinase
Erratum in
-
Molecular characterization of a mammalian smooth muscles myosin light chain kinase.J Biol Chem. 1992 May 5;267(13):9450. J Biol Chem. 1992. PMID: 1577772 No abstract available.
Abstract
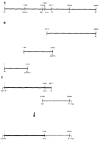
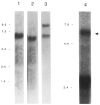
A 5.6-kilobase cDNA clone has been isolated which includes the entire coding region for the myosin light chain kinase from rabbit uterine tissue. This cDNA, expressed in COS cells, encodes a Ca2+/calmodulin-dependent protein kinase with catalytic properties similar to other purified smooth muscle myosin light chain kinases. A module (TLKPVGNIKPAE), repeated sequentially 15 times, has been identified near the N terminus of this smooth muscle kinase. It is not present in chicken gizzard or rabbit skeletal muscle myosin light chain kinases. This repeat module and a subrepeat (K P A/V) are similar in amino acid content to repeated motifs present in other proteins, some of which have been shown to associate with chromatin structures. Immunoblot analysis after sodium dodecyl sulfate-polyacrylamide gel electrophoresis, used to compare myosin light chain kinase present in rabbit, bovine, and chicken smooth and nonmuscle tissues, showed that within each species both tissue types have myosin light chain kinases with indistinguishable molecular masses. These data suggest that myosin light chain kinases present in smooth and nonmuscle tissues are the same protein.
Figures





Similar articles
-
Isolation of cDNA for bovine stomach 155 kDa protein exhibiting myosin light chain kinase activity.J Biochem. 1992 Dec;112(6):786-91. doi: 10.1093/oxfordjournals.jbchem.a123976. J Biochem. 1992. PMID: 1284247
-
Regulatory and structural motifs of chicken gizzard myosin light chain kinase.Proc Natl Acad Sci U S A. 1990 Mar;87(6):2284-8. doi: 10.1073/pnas.87.6.2284. Proc Natl Acad Sci U S A. 1990. PMID: 2315320 Free PMC article.
-
The carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein, telokin.J Biol Chem. 1991 Dec 15;266(35):23945-52. J Biol Chem. 1991. PMID: 1748667 Free PMC article.
-
Phosphorylation of myosin light chain kinase: a cellular mechanism for Ca2+ desensitization.Mol Cell Biochem. 1993 Nov;127-128:229-37. doi: 10.1007/BF01076774. Mol Cell Biochem. 1993. PMID: 7935354 Review.
-
Myosin light chain kinase phosphorylation: regulation of the Ca2+ sensitivity of contractile elements.Adv Exp Med Biol. 1991;304:129-38. doi: 10.1007/978-1-4684-6003-2_12. Adv Exp Med Biol. 1991. PMID: 1803895 Review.
Cited by
-
Myosin light chain kinases.J Muscle Res Cell Motil. 1997 Feb;18(1):1-16. doi: 10.1023/a:1018616814417. J Muscle Res Cell Motil. 1997. PMID: 9147985 Review. No abstract available.
-
Myosin light chain kinases: division of work in cell migration.Cell Adh Migr. 2009 Jul-Sep;3(3):256-8. doi: 10.4161/cam.3.3.8212. Epub 2009 Jul 17. Cell Adh Migr. 2009. PMID: 19276674 Free PMC article.
-
INSIGHTS INTO THE ROLES OF NON-MUSCLE MYOSIN IIA IN HUMAN KERATINOCYTE MIGRATION.Cell Mol Bioeng. 2009 Nov 21;2(4):486-494. doi: 10.1007/s12195-009-0094-2. Cell Mol Bioeng. 2009. PMID: 20548965 Free PMC article.
-
New nucleotide sequence data on the EMBL File Server.Nucleic Acids Res. 1992 Oct 25;20(20):5495-515. doi: 10.1093/nar/20.20.5495. Nucleic Acids Res. 1992. PMID: 1437578 Free PMC article. No abstract available.
-
Role of the short isoform of myosin light chain kinase in the contraction of cultured smooth muscle cells as examined by its down-regulation.Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9556-61. doi: 10.1073/pnas.142298599. Epub 2002 Jun 26. Proc Natl Acad Sci U S A. 2002. PMID: 12087128 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

