The carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein, telokin
- PMID: 1748667
- PMCID: PMC2836763
The carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein, telokin
Abstract
It has been proposed that the carboxyl terminus of the smooth muscle myosin light chain kinase is expressed as an independent protein. This protein has been purified from tissues and named telokin (Ito, M., Dabrowska, R., Guerriero, V., Jr., and Hartshorne, D. J. (1989) J. Biol. Chem. 264, 13971-13974). In this study we have isolated and characterized cDNA and genomic clones encoding telokin. Analysis of a genomic DNA clone suggests that the mRNA encoding telokin arises from a promoter which appears to be located within an intron of the smooth muscle myosin light chain kinase (MLCK) gene. This intron interrupts exons encoding the calmodulin binding domain of the kinase. The amino acid sequence deduced from the cDNA predicts that telokin is identical to the carboxyl-terminal 155 residues of the smooth muscle MLCK. Unlike the smooth muscle MLCK which is expressed in both smooth and non-muscle tissues, telokin is expressed in some smooth muscle tissues but has not been detected in aortic smooth muscle or in any non-muscle tissues.
Figures


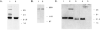

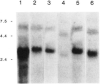


References
-
- Adelstein RS, Conti MA, Daniel JL, Anderson W., Jr. Biochemistry and Pharmacology of Platelets. Elsevier Science Publishing Co.; North Holland, The Netherlands: 1975. pp. 101–119.
-
- Adelstein RS, Conti MA, Hathaway DR, Klee CB. J. Biol. Chem. 1978;253:8347–8350. - PubMed
-
- Andersson S, Davis DL, Dahlbäch H, Jörnvall H, Russell DW. J. Biol. Chem. 1989;264:8222–8229. - PubMed
-
- Arfin SM, Bradshaw RA. Biochemistry. 1988;27:7979–7984. - PubMed
-
- Bagchi IC, Kemp BE, Means AR. J. Biol. Chem. 1989;264:15843–15849. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

