Mutational analysis of EGFR and related signaling pathway genes in lung adenocarcinomas identifies a novel somatic kinase domain mutation in FGFR4
- PMID: 17487277
- PMCID: PMC1855985
- DOI: 10.1371/journal.pone.0000426
Mutational analysis of EGFR and related signaling pathway genes in lung adenocarcinomas identifies a novel somatic kinase domain mutation in FGFR4
Abstract
Background: Fifty percent of lung adenocarcinomas harbor somatic mutations in six genes that encode proteins in the EGFR signaling pathway, i.e., EGFR, HER2/ERBB2, HER4/ERBB4, PIK3CA, BRAF, and KRAS. We performed mutational profiling of a large cohort of lung adenocarcinomas to uncover other potential somatic mutations in genes of this signaling pathway that could contribute to lung tumorigenesis.
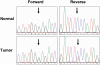
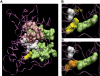
Methodology/principal findings: We analyzed genomic DNA from a total of 261 resected, clinically annotated non-small cell lung cancer (NSCLC) specimens. The coding sequences of 39 genes were screened for somatic mutations via high-throughput dideoxynucleotide sequencing of PCR-amplified gene products. Mutations were considered to be somatic only if they were found in an independent tumor-derived PCR product but not in matched normal tissue. Sequencing of 9MB of tumor sequence identified 239 putative genetic variants. We further examined 22 variants found in RAS family genes and 135 variants localized to exons encoding the kinase domain of respective proteins. We identified a total of 37 non-synonymous somatic mutations; 36 were found collectively in EGFR, KRAS, BRAF, and PIK3CA. One somatic mutation was a previously unreported mutation in the kinase domain (exon 16) of FGFR4 (Glu681Lys), identified in 1 of 158 tumors. The FGFR4 mutation is analogous to a reported tumor-specific somatic mutation in ERBB2 and is located in the same exon as a previously reported kinase domain mutation in FGFR4 (Pro712Thr) in a lung adenocarcinoma cell line.
Conclusions/significance: This study is one of the first comprehensive mutational analyses of major genes in a specific signaling pathway in a sizeable cohort of lung adenocarcinomas. Our results suggest the majority of gain-of-function mutations within kinase genes in the EGFR signaling pathway have already been identified. Our findings also implicate FGFR4 in the pathogenesis of a subset of lung adenocarcinomas.
Conflict of interest statement
Figures

References
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. - PubMed
-
- Gabrielson E. Worldwide trends in lung cancer pathology. Respirology. 2006;11:533–538. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
-
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

