Probing Na(+)-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: new insights into the role of counterions and electrostatic interactions in adaptive recognition
- PMID: 17488097
- PMCID: PMC3319146
- DOI: 10.1021/bi700335n
Probing Na(+)-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: new insights into the role of counterions and electrostatic interactions in adaptive recognition
Abstract
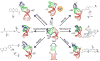
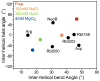
Many regulatory RNAs undergo large changes in structure upon recognition of proteins and ligands, but the mechanism by which this occurs remains poorly understood. Using NMR residual dipolar coupling (RDCs), we characterized Na+-induced changes in the structure and dynamics of the bulge-containing HIV-1 transactivation response element (TAR) RNA that mirrors changes induced by small molecules bearing a different number of cationic groups. Increasing the Na+ concentration from 25 to 320 mM led to a continuous reduction in the average inter-helical bend angle (from 46 degrees to 22 degrees ), inter-helical twist angle (from 66 degrees to -18 degrees ), and inter-helix flexibility (as measured by an increase in the internal generalized degree of order from 0.56 to 0.74). Similar conformational changes were observed with Mg2+, indicating that nonspecific electrostatic interactions drive the conformational transition, although results also suggest that Na+ and Mg2+ may associate with TAR in distinct modes. The transition can be rationalized on the basis of a population-weighted average of two ensembles comprising an electrostatically relaxed bent and flexible TAR conformation that is weakly associated with counterions and a globally rigid coaxial conformation that has stronger electrostatic potential and association with counterions. The TAR inter-helical orientations that are stabilized by small molecules fall around the metal-induced conformational pathway, indicating that counterions may help predispose the TAR conformation for target recognition. Our results underscore the intricate sensitivity of RNA conformational dynamics to environmental conditions and demonstrate the ability to detect subtle conformational changes using NMR RDCs.
Figures





References
-
- Leulliot N, Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. - PubMed
-
- Micura R, Hobartner C. On secondary structure rearrangements and equilibria of small RNAs. Chembiochem. 2003;4:984–990. - PubMed
-
- Al-Hashimi HM. Dynamics-Based Amplification of RNA Function and Its Characterization by Using NMR Spectroscopy. Chembiochem. 2005;6:1506–1519. - PubMed
-
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. - PubMed
-
- Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

