Characterization of molecular recognition features, MoRFs, and their binding partners
- PMID: 17488107
- PMCID: PMC2570643
- DOI: 10.1021/pr0701411
Characterization of molecular recognition features, MoRFs, and their binding partners
Abstract
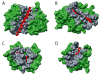
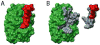
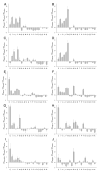
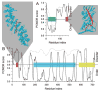
Molecular Recognition Features (MoRFs) are short, interaction-prone segments of protein disorder that undergo disorder-to-order transitions upon specific binding, representing a specific class of intrinsically disordered regions that exhibit molecular recognition and binding functions. MoRFs are common in various proteomes and occupy a unique structural and functional niche in which function is a direct consequence of intrinsic disorder. Example MoRFs collected from the Protein Data Bank (PDB) have been divided into three subtypes according to their structures in the bound state: alpha-MoRFs form alpha-helices, beta-MoRFs form beta-strands, and iota-MoRFs form structures without a regular pattern of backbone hydrogen bonds. These example MoRFs were indicated to be intrinsically disordered in the absence of their binding partners by several criteria. In this study, we used several geometric and physiochemical criteria to examine the properties of 62 alpha-, 20 beta-, and 176 iota-MoRF complex structures. Interface residues were examined by calculating differences in accessible surface area between the complex and isolated monomers. The compositions and physiochemical properties of MoRF and MoRF partner interface residues were compared to the interface residues of homodimers, heterodimers, and antigen-antibody complexes. Our analysis indicates that there are significant differences in residue composition and several geometric and physicochemical properties that can be used to discriminate, with a high degree of accuracy, between various interfaces in protein interaction data sets. Implications of these findings for the development of MoRF-partner interaction predictors are discussed. In addition, structural changes upon MoRF-to-partner complex formation were examined for several illustrative examples.
Figures


References
-
- Argos P. An investigation of protein subunit and domain interfaces. Protein Eng. 1988;2:101–113. - PubMed
-
- Chothia C, Janin J. Principles of protein-protein recognition. Nature. 1975;256:705–708. - PubMed
-
- Janin J, Chothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990;265:16027–16030. - PubMed
-
- Korn AP, Burnett RM. Distribution and complementarity of hydropathy in multisubunit proteins. Proteins. 1991;9:37–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

