Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5' and 3' end formation
- PMID: 17488843
- PMCID: PMC1920247
- DOI: 10.1093/nar/gkm270
Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5' and 3' end formation
Abstract
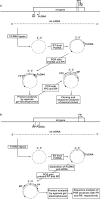
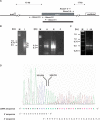
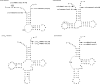
With CR-RT-PCR as primary approach we mapped the 5' and 3' transcript ends of all mitochondrial protein-coding genes in Arabidopsis thaliana. Almost all transcripts analyzed have single major 3' termini, while multiple 5' ends were found for several genes. Some of the identified 5' ends map within promoter motifs suggesting these ends to be derived from transcription initiation while the majority of the 5' termini seems to be generated post-transcriptionally. Assignment of the extremities of 5' leader RNAs revealed clear evidence for an endonucleolytic generation of the major cox1 and atp9 5' mRNA ends. tRNA-like structures, so-called t-elements, are associated either with 5' or with 3' termini of several mRNAs. These secondary structures most likely act as cis-signals for endonucleolytic cleavages by RNase Z and/or RNase P. Since no conserved sequence motif is evident at post-transcriptionally derived ends, we suggest t-elements, stem-loops and probably complex higher order structures as cis-elements for processing. This analysis provides novel insights into 5' and 3' end formation of mRNAs. In addition, the complete transcript map is a substantial and important basis for future studies of gene expression in mitochondria of higher plants.
Figures

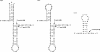
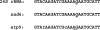

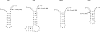
Similar articles
-
RT-PCR analysis of 5' to 3'-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria.Nucleic Acids Res. 2002 Jan 15;30(2):439-46. doi: 10.1093/nar/30.2.439. Nucleic Acids Res. 2002. PMID: 11788705 Free PMC article.
-
RNA Processing Factor 5 is required for efficient 5' cleavage at a processing site conserved in RNAs of three different mitochondrial genes in Arabidopsis thaliana.Plant J. 2013 May;74(4):593-604. doi: 10.1111/tpj.12143. Epub 2013 Apr 15. Plant J. 2013. PMID: 23398165
-
3'-Inverted repeats in plant mitochondrial mRNAs are processing signals rather than transcription terminators.EMBO J. 1997 Aug 15;16(16):5069-76. doi: 10.1093/emboj/16.16.5069. EMBO J. 1997. PMID: 9305648 Free PMC article.
-
Maturation of 5' ends of plant mitochondrial RNAs.Physiol Plant. 2016 Jul;157(3):280-8. doi: 10.1111/ppl.12423. Epub 2016 Mar 23. Physiol Plant. 2016. PMID: 26833432 Review.
-
P-class pentatricopeptide repeat proteins are required for efficient 5' end formation of plant mitochondrial transcripts.RNA Biol. 2013;10(9):1511-9. doi: 10.4161/rna.26129. Epub 2013 Aug 15. RNA Biol. 2013. PMID: 24184847 Free PMC article. Review.
Cited by
-
A maternally inherited DNA marker, descended from Solanum demissum (2n = 6x = 72) to S. tuberosum (2n = 4x = 48).Breed Sci. 2011 Dec;61(4):426-34. doi: 10.1270/jsbbs.61.426. Epub 2011 Dec 15. Breed Sci. 2011. PMID: 23136481 Free PMC article.
-
Sorting of mitochondrial and plastid heteroplasmy in Arabidopsis is extremely rapid and depends on MSH1 activity.Proc Natl Acad Sci U S A. 2022 Aug 23;119(34):e2206973119. doi: 10.1073/pnas.2206973119. Epub 2022 Aug 15. Proc Natl Acad Sci U S A. 2022. PMID: 35969753 Free PMC article.
-
Major contribution of transcription initiation to 5'-end formation of mitochondrial steady-state transcripts in maize.RNA Biol. 2019 Jan;16(1):104-117. doi: 10.1080/15476286.2018.1561604. Epub 2019 Jan 6. RNA Biol. 2019. PMID: 30585757 Free PMC article.
-
Mitochondrial mRNA polymorphisms in different Arabidopsis accessions.Plant Physiol. 2008 Oct;148(2):1106-16. doi: 10.1104/pp.108.126201. Epub 2008 Aug 6. Plant Physiol. 2008. PMID: 18685042 Free PMC article.
-
Rapid Shifts in Mitochondrial tRNA Import in a Plant Lineage with Extensive Mitochondrial tRNA Gene Loss.Mol Biol Evol. 2021 Dec 9;38(12):5735-5751. doi: 10.1093/molbev/msab255. Mol Biol Evol. 2021. PMID: 34436590 Free PMC article.
References
-
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics. 2002;268:434–445. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

