e-Science and biological pathway semantics
- PMID: 17493286
- PMCID: PMC1892100
- DOI: 10.1186/1471-2105-8-S3-S3
e-Science and biological pathway semantics
Abstract
Background: The development of e-Science presents a major set of opportunities and challenges for the future progress of biological and life scientific research. Major new tools are required and corresponding demands are placed on the high-throughput data generated and used in these processes. Nowhere is the demand greater than in the semantic integration of these data. Semantic Web tools and technologies afford the chance to achieve this semantic integration. Since pathway knowledge is central to much of the scientific research today it is a good test-bed for semantic integration. Within the context of biological pathways, the BioPAX initiative, part of a broader movement towards the standardization and integration of life science databases, forms a necessary prerequisite for its successful application of e-Science in health care and life science research. This paper examines whether BioPAX, an effort to overcome the barrier of disparate and heterogeneous pathway data sources, addresses the needs of e-Science.
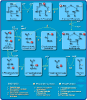
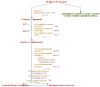
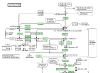
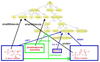
Results: We demonstrate how BioPAX pathway data can be used to ask and answer some useful biological questions. We find that BioPAX comes close to meeting a broad range of e-Science needs, but certain semantic weaknesses mean that these goals are missed. We make a series of recommendations for re-modeling some aspects of BioPAX to better meet these needs.
Conclusion: Once these semantic weaknesses are addressed, it will be possible to integrate pathway information in a manner that would be useful in e-Science.
Figures


References
-
- Hey T, Trefethen A. e-Science and its implications. Philos Transact A Math Phys Eng Sci. 2003;361:1809–1825. http://dx.doi.org/10.1098/rsta.2003.1224 - DOI - PubMed
-
- Hey T, Trefethen AE. Cyberinfrastructure for e-Science. Science. 2005;308:817–821. http://dx.doi.org/10.1126/science.1110410 - DOI - PubMed
-
- Goble C, Pettifer S, Stevens R, Greenhalgh C. Knowledge Integration: In Silico Experiments in Bioinformatics. In: Foster I, Kesselman C, editor. The Grid: Blueprint for a New Computing Infrastructure. second. Morgan Kaufmann Publishers; 2003. pp. 121–134.
-
- Stein L. Creating a bioinformatics nation. Nature. 2002;417:119–120. http://dx.doi.org/10.1038/417119a - DOI - PubMed
-
- Goble C, Wroe C, Stevens R, the myGrid consortium The myGrid Project: Services, Architecture and Demonstrator. Proc UK e-Science programme All Hands Conference. 2003. pp. 595–603.http://www.cs.man.ac.uk/~stevensr/papers/all-hands-general.pdf
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

