Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment
- PMID: 17494080
- PMCID: PMC1933369
- DOI: 10.1128/JVI.00083-07
Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment
Abstract
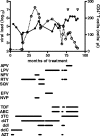
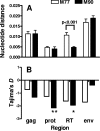
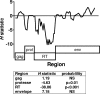
Viral recombination has been postulated to play two roles in the development of human immunodeficiency virus (HIV) resistance to antiretroviral drugs. First, recombination has the capacity to associate resistance mutations expressed by distinct viruses, thereby contributing to the development of viruses with improved drug resistance. In addition, recombination could preserve diversity in regions outside those subject to strong selective pressure. In this study, we sought direct evidence for the occurrence of these processes in vivo by evaluating clonal virus populations obtained from the same patient before and after a treatment change that, while unsuccessful in controlling viral replication, led to the emergence of viruses expressing a different profile of resistance mutations. Phylogenetic studies supported the conclusion that the genotype arising after the treatment change resulted from the emergence of recombinant viruses carrying previously existing resistance mutations in novel combinations, whereas alternative explanations, including convergent evolution, were not consistent with observed genotypic changes. Despite evidence for a strong loss of genetic diversity in genomic regions coding for the protease and reverse transcriptase, diversity in regions coding for Gag and envelope was considerably higher, and recombination between the emerging viruses expressing the new pattern of resistance mutations and viral quasispecies in the previously dominant population contributed to this preservation of diversity in the envelope gene. These findings emphasize that recombination can participate in the adaptation of HIV to changing selective pressure, both by generating novel combinations of resistance mutations and by maintaining diversity in genomic regions outside those implicated in a selective sweep.
Figures


References
-
- Barbour, J. D., F. M. Hecht, T. Wrin, T. J. Liegler, C. A. Ramstead, M. P. Busch, M. R. Segal, C. J. Petropoulos, and R. M. Grant. 2004. Persistence of primary drug resistance among recently HIV-1 infected adults. AIDS 18:1683-1689. - PubMed
-
- Brenner, B., J. P. Routy, Y. Quan, D. Moisi, M. Oliveira, D. Turner, and M. A. Wainberg. 2004. Persistence of multidrug-resistant HIV-1 in primary infection leading to superinfection. AIDS 18:1653-1660. - PubMed
-
- Brown, A. J., and A. Cleland. 1996. Independent evolution of the env and pol genes of HIV-1 during zidovudine therapy. AIDS 10:1067-1073. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

