Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast
- PMID: 17494733
- PMCID: PMC1885633
- DOI: 10.1073/pnas.0703162104
Evidence for regulatory function of nucleus-encoded factors on mRNA stabilization and translation in the chloroplast
Abstract
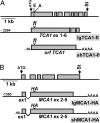
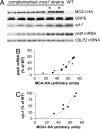
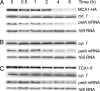
A salient feature of organelle gene expression is the requirement for nucleus-encoded factors that act posttranscriptionally in a gene-specific manner. A central issue is to understand whether these factors are merely constitutive or have a regulatory function. In the unicellular alga Chlamydomonas reinhardtii, expression of the chloroplast petA gene-encoding cytochrome f, a major subunit of the cytochrome b(6)f complex, depends on two specific nucleus-encoded factors: MCA1, required for stable accumulation of the petA transcript, and TCA1, required for its translation. We cloned the TCA1 gene, encoding a pioneer protein, and transformed appropriate mutant strains with tagged versions of MCA1 and TCA1. In transformed strains expressing decreasing amounts of MCA1 or TCA1, the concentration of these factors proved limiting for petA mRNA accumulation and cytochrome f translation, respectively. This observation suggests that in exponentially growing cells, the abundance of MCA1 sets the pool of petA transcripts, some of which are TCA1-selected for an assembly-dependent translation of cytochrome f. We show that MCA1 is a short-lived protein. Its abundance varies rapidly with physiological conditions that deeply affect expression of the petA gene in vivo, for instance in aging cultures or upon changes in nitrogen availability. We observed similar but more limited changes in the abundance of TCA1. We conclude that in conditions where de novo biogenesis of cytochrome b(6)f complexes is not required, a rapid drop in MCA1 exhausts the pool of petA transcripts, and the progressive loss of TCA1 further prevents translation of cytochrome f.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


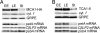

Similar articles
-
Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts.Mol Cell Biol. 2008 Sep;28(17):5529-42. doi: 10.1128/MCB.02056-07. Epub 2008 Jun 23. Mol Cell Biol. 2008. PMID: 18573878 Free PMC article.
-
TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast.Genetics. 2001 Sep;159(1):119-32. doi: 10.1093/genetics/159.1.119. Genetics. 2001. PMID: 11560891 Free PMC article.
-
The nucleus-encoded trans-acting factor MCA1 plays a critical role in the regulation of cytochrome f synthesis in Chlamydomonas chloroplasts.Plant Cell. 2011 Jan;23(1):333-49. doi: 10.1105/tpc.110.078170. Epub 2011 Jan 7. Plant Cell. 2011. PMID: 21216944 Free PMC article.
-
Assembly-controlled regulation of chloroplast gene translation.Biochem Soc Trans. 2001 Aug;29(Pt 4):421-6. doi: 10.1042/bst0290421. Biochem Soc Trans. 2001. PMID: 11498001 Review.
-
The unicellular green alga Chlamydomonas reinhardtii as an experimental system to study chloroplast RNA metabolism.Naturwissenschaften. 2000 Mar;87(3):97-107. doi: 10.1007/s001140050686. Naturwissenschaften. 2000. PMID: 10798194 Review.
Cited by
-
Coordination of gene expression between organellar and nuclear genomes.Nat Rev Genet. 2008 May;9(5):383-95. doi: 10.1038/nrg2348. Nat Rev Genet. 2008. PMID: 18368053 Free PMC article. Review.
-
Molecular basis of fruit development.Front Plant Sci. 2015 Feb 5;6:28. doi: 10.3389/fpls.2015.00028. eCollection 2015. Front Plant Sci. 2015. PMID: 25699063 Free PMC article. No abstract available.
-
High intraspecific diversity of Restorer-of-fertility-like genes in barley.Plant J. 2019 Jan;97(2):281-295. doi: 10.1111/tpj.14115. Epub 2018 Nov 9. Plant J. 2019. PMID: 30276910 Free PMC article.
-
Pas de Trois: An Overview of Penta-, Tetra-, and Octo-Tricopeptide Repeat Proteins From Chlamydomonas reinhardtii and Their Role in Chloroplast Gene Expression.Front Plant Sci. 2021 Nov 17;12:775366. doi: 10.3389/fpls.2021.775366. eCollection 2021. Front Plant Sci. 2021. PMID: 34868174 Free PMC article. Review.
-
Small RNAs reveal two target sites of the RNA-maturation factor Mbb1 in the chloroplast of Chlamydomonas.Nucleic Acids Res. 2014 Mar;42(5):3286-97. doi: 10.1093/nar/gkt1272. Epub 2013 Dec 11. Nucleic Acids Res. 2014. PMID: 24335082 Free PMC article.
References
-
- Barkan A, Goldschmidt-Clermont M. Biochimie. 2000;82:559–572. - PubMed
-
- Fox TD. Genetics of Mitochondrial Translation. Cold Spring Harbor, New York: Cold Spring Harbor Lab Press; 1996.
-
- Gillham NW, Boynton JE, Hauser CR. Annu Rev Genet. 1994;28:71–93. - PubMed
-
- Grivell LA. Crit Rev Biochem Mol Biol. 1995;30:121–164. - PubMed
-
- Monde RA, Schuster G, Stern DB. Biochimie. 2000;82:573–582. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

