PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal
- PMID: 17494868
- PMCID: PMC1924815
- DOI: 10.1091/mbc.e06-10-0897
PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal
Abstract
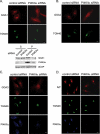
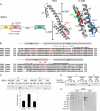
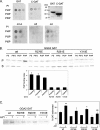
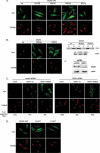
Phosphatidylinositol 4 phosphate (PI4P) is highly enriched in the trans-Golgi network (TGN). Here we establish that PI4P is a key regulator of the recruitment of the GGA clathrin adaptor proteins to the TGN and that PI4P has a novel role in promoting their recognition of the ubiquitin (Ub) sorting signal. Knockdown of PI4KIIalpha by RNA interference (RNAi), which depletes the TGN's PI4P, impaired the recruitment of the GGAs to the TGN. GGAs bind PI4P primarily through their GAT domain, in a region called C-GAT, which also binds Ub but not Arf1. We identified two basic residues in the GAT domain that are essential for PI4P binding in vitro and for the recruitment of GGAs to the TGN in vivo. Unlike wild-type GGA, GGA with mutated GATs failed to rescue the abnormal TGN phenotype of the GGA RNAi-depleted cells. These residues partially overlap with those that bind Ub, and PI4P increased the affinity of the GAT domain for Ub. Because the recruitment of clathrin adaptors and their cargoes to the TGN is mediated through a web of low-affinity interactions, our results show that the dual roles of PI4P can promote specific GGA targeting and cargo recognition at the TGN.
Figures
Similar articles
-
The GAT domains of clathrin-associated GGA proteins have two ubiquitin binding motifs.J Biol Chem. 2004 Dec 24;279(52):54808-16. doi: 10.1074/jbc.M406654200. Epub 2004 Oct 19. J Biol Chem. 2004. PMID: 15494413 Free PMC article.
-
Molecular mechanism of membrane recruitment of GGA by ARF in lysosomal protein transport.Nat Struct Biol. 2003 May;10(5):386-93. doi: 10.1038/nsb920. Nat Struct Biol. 2003. PMID: 12679809
-
Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, gamma-ear-containing, ADP-ribosylation-factor-binding proteins.Proc Natl Acad Sci U S A. 2005 Feb 15;102(7):2334-9. doi: 10.1073/pnas.0500118102. Epub 2005 Feb 8. Proc Natl Acad Sci U S A. 2005. PMID: 15701688 Free PMC article.
-
Cargo Sorting at the trans-Golgi Network for Shunting into Specific Transport Routes: Role of Arf Small G Proteins and Adaptor Complexes.Cells. 2019 Jun 3;8(6):531. doi: 10.3390/cells8060531. Cells. 2019. PMID: 31163688 Free PMC article. Review.
-
The structure and function of GGAs, the traffic controllers at the TGN sorting crossroads.Cell Struct Funct. 2003 Oct;28(5):431-42. doi: 10.1247/csf.28.431. Cell Struct Funct. 2003. PMID: 14745135 Review.
Cited by
-
Sec14 like PITPs couple lipid metabolism with phosphoinositide synthesis to regulate Golgi functionality.Subcell Biochem. 2012;59:271-87. doi: 10.1007/978-94-007-3015-1_9. Subcell Biochem. 2012. PMID: 22374094 Free PMC article. Review.
-
Phosphatidylinositol 4,5-bisphosphate controls Rab7 and PLEKHM1 membrane cycling during autophagosome-lysosome fusion.EMBO J. 2019 Apr 15;38(8):e100312. doi: 10.15252/embj.2018100312. Epub 2019 Mar 13. EMBO J. 2019. PMID: 31368593 Free PMC article.
-
Pi4KIIα Regulates Unconditioned Stimulus-Retrieval-Induced Fear Memory Reconsolidation through Endosomal Trafficking of AMPA Receptors.iScience. 2020 Mar 27;23(3):100895. doi: 10.1016/j.isci.2020.100895. Epub 2020 Feb 8. iScience. 2020. PMID: 32088394 Free PMC article.
-
Chromosomal Instability and Phosphoinositide Pathway Gene Signatures in Glioblastoma Multiforme.Mol Neurobiol. 2016 Jan;53(1):621-630. doi: 10.1007/s12035-014-9034-9. Epub 2014 Dec 15. Mol Neurobiol. 2016. PMID: 25502460 Free PMC article.
-
Tip growth: signaling in the apical dome.Curr Opin Plant Biol. 2008 Dec;11(6):662-71. doi: 10.1016/j.pbi.2008.10.002. Epub 2008 Oct 30. Curr Opin Plant Biol. 2008. PMID: 18977167 Free PMC article. Review.
References
-
- Akutsu M., Kawasaki M., Katoh Y., Shiba T., Yamaguchi Y., Kato R., Kato K., Nakayama K., Wakatsuki S. Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain. FEBS Lett. 2005;579:5385–5391. - PubMed
-
- Barylko B., Gerber S. H., Binns D. D., Grichine N., Khvotchev M., Sudhof T. C., Albanesi J. P. A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J. Biol. Chem. 2001;276:7705–7708. - PubMed
-
- Behnia R., Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. - PubMed
-
- Bonifacino J. S. The GGA proteins: adaptors on the move. Nat. Rev. Mol. Cell Biol. 2004;5:23–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

