Evolutionarily conserved genes preferentially accumulate introns
- PMID: 17495009
- PMCID: PMC1899115
- DOI: 10.1101/gr.5978207
Evolutionarily conserved genes preferentially accumulate introns
Abstract
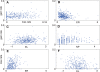
Introns that interrupt eukaryotic protein-coding sequences are generally thought to be nonfunctional. However, for reasons still poorly understood, positions of many introns are highly conserved in evolution. Previous reconstructions of intron gain and loss events during eukaryotic evolution used a variety of simplified evolutionary models that yielded contradicting conclusions and are not suited to reveal some of the key underlying processes. We combine a comprehensive probabilistic model and an extended data set, including 391 conserved genes from 19 eukaryotes, to uncover previously unnoticed aspects of intron evolution--in particular, to assign intron gain and loss rates to individual genes. The rates of intron gain and loss in a gene show moderate positive correlation. A gene's intron gain rate shows a highly significant negative correlation with the coding-sequence evolution rate; intron loss rate also significantly, but positively, correlates with the sequence evolution rate. Correlations of the opposite signs, albeit less significant ones, are observed between intron gain and loss rates and gene expression level. It is proposed that intron evolution includes a neutral component, which is manifest in the positive correlation between the gain and loss rates and a selection-driven component as reflected in the links between intron gain and loss and sequence evolution. The increased intron gain and decreased intron loss in evolutionarily conserved genes indicate that intron insertion often might be adaptive, whereas some of the intron losses might be deleterious. This apparent functional importance of introns is likely to be due, at least in part, to their multiple effects on gene expression.
Figures
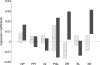
Similar articles
-
Origin and evolution of spliceosomal introns.Biol Direct. 2012 Apr 16;7:11. doi: 10.1186/1745-6150-7-11. Biol Direct. 2012. PMID: 22507701 Free PMC article. Review.
-
Analysis of evolution of exon-intron structure of eukaryotic genes.Brief Bioinform. 2005 Jun;6(2):118-34. doi: 10.1093/bib/6.2.118. Brief Bioinform. 2005. PMID: 15975222 Review.
-
Patterns of intron gain and conservation in eukaryotic genes.BMC Evol Biol. 2007 Oct 12;7:192. doi: 10.1186/1471-2148-7-192. BMC Evol Biol. 2007. PMID: 17935625 Free PMC article.
-
Spliceosomal Introns: Features, Functions, and Evolution.Biochemistry (Mosc). 2020 Jul;85(7):725-734. doi: 10.1134/S0006297920070019. Biochemistry (Mosc). 2020. PMID: 33040717 Review.
-
How Common Is Parallel Intron Gain? Rapid Evolution Versus Independent Creation in Recently Created Introns in Daphnia.Mol Biol Evol. 2016 Aug;33(8):1902-6. doi: 10.1093/molbev/msw091. Epub 2016 May 10. Mol Biol Evol. 2016. PMID: 27189562
Cited by
-
Intron presence-absence polymorphisms in Daphnia.Mol Biol Evol. 2008 Oct;25(10):2129-39. doi: 10.1093/molbev/msn164. Epub 2008 Jul 29. Mol Biol Evol. 2008. PMID: 18667441 Free PMC article.
-
Evolution of GHF5 endoglucanase gene structure in plant-parasitic nematodes: no evidence for an early domain shuffling event.BMC Evol Biol. 2008 Nov 3;8:305. doi: 10.1186/1471-2148-8-305. BMC Evol Biol. 2008. PMID: 18980666 Free PMC article.
-
Genes with a large intronic burden show greater evolutionary conservation on the protein level.BMC Evol Biol. 2014 Mar 16;14(1):50. doi: 10.1186/1471-2148-14-50. BMC Evol Biol. 2014. PMID: 24629165 Free PMC article.
-
Effects of DNA Methylation and Chromatin State on Rates of Molecular Evolution in Insects.G3 (Bethesda). 2015 Dec 4;6(2):357-63. doi: 10.1534/g3.115.023499. G3 (Bethesda). 2015. PMID: 26637432 Free PMC article.
-
Identifying the mechanisms of intron gain: progress and trends.Biol Direct. 2012 Sep 10;7:29. doi: 10.1186/1745-6150-7-29. Biol Direct. 2012. PMID: 22963364 Free PMC article. Review.
References
-
- Castillo-Davis C.I., Bedford T.B., Hartl D.L., Bedford T.B., Hartl D.L., Hartl D.L. Accelerated rates of intron gain/loss and protein evolution in duplicate genes in human and mouse malaria parasites. Mol. Biol. Evol. 2004;21:1422–1427. - PubMed
-
- Collins L., Penny D., Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol. Biol. Evol. 2005;22:1053–1066. - PubMed
-
- Csuros M. Likely scenarios of intron evolution. Comparative Genomics. Lecture Notes in Computer Science. 2005;3678:47–60.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
