Identification of genomic classifiers that distinguish induction failure in T-lineage acute lymphoblastic leukemia: a report from the Children's Oncology Group
- PMID: 17495134
- PMCID: PMC1975833
- DOI: 10.1182/blood-2006-12-059790
Identification of genomic classifiers that distinguish induction failure in T-lineage acute lymphoblastic leukemia: a report from the Children's Oncology Group
Erratum in
- Blood. 2008 May 1;111(9):4830
Abstract
The clinical and cytogenetic features associated with T-cell acute lymphoblastic leukemia (T-ALL) are not predictive of early treatment failure. Based on the hypothesis that microarrays might identify patients who fail therapy, we used the Affymetrix U133 Plus 2.0 chip and prediction analysis of microarrays (PAM) to profile 50 newly diagnosed patients who were treated in the Children's Oncology Group (COG) T-ALL Study 9404. We identified a 116-member genomic classifier that could accurately distinguish all 6 induction failure (IF) cases from 44 patients who achieved remission; network analyses suggest a prominent role for genes mediating cellular quiescence. Seven genes were similarly upregulated in both the genomic classifier for IF patients and T-ALL cell lines having acquired resistance to neoplastic agents, identifying potential target genes for further study in drug resistance. We tested whether our classifier could predict IF within 42 patient samples obtained from COG 8704 and, using PAM to define a smaller classifier for the U133A chip, correctly identified the single IF case and patients with persistently circulating blasts. Genetic profiling may identify T-ALL patients who are likely to fail induction and for whom alternate treatment strategies might be beneficial.
Figures
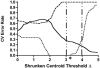

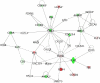
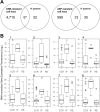
References
-
- Amylon MD, Shuster J, Pullen J, et al. Intensive high-dose asparaginase consolidation improves survival for pediatric patients with T cell acute lymphoblastic leukemia and advanced stage lymphoblastic lymphoma: a Pediatric Oncology Group study. Leukemia. 1999;13:335–342. - PubMed
-
- Pullen J, Shuster JJ, Link MP, et al. Significance of commonly used prognostic factors differs for children with T-cell acute lymphoblastic leukemia (ALL) compared with those with B-precursor ALL: a Pediatric Oncology Group Study. Leukemia. 1999;13:1696–1707. - PubMed
-
- Uckun FM, Sensel MG, Sun L, et al. Biology and treatment of childhood T-Lineage acute lymphoblastic leukemia. Blood. 1998;91:735–746. - PubMed
-
- Shuster JJ, Falletta JM, Pullen DJ, et al. Prognostic factors in childhood T-cell acute lymphoblastic leukemia: a Pediatric Oncology Group Study. Blood. 1990;75:166–173. - PubMed
-
- Winick NJ, Carroll WL, Hunger SP. Childhood leukemia: new advances and challenges. N Engl J Med. 2004;351:601–603. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

