Step length measurement--theory and simulation for tethered bead constant-force single molecule assay
- PMID: 17496045
- PMCID: PMC1913155
- DOI: 10.1529/biophysj.106.097915
Step length measurement--theory and simulation for tethered bead constant-force single molecule assay
Abstract
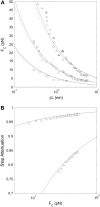
Linear molecular motors translocate along polymeric tracks using discrete steps. The step length is usually measured using constant-force single molecule experiments in which the polymer is tethered to a force-clamped microsphere. During the enzymatic cycle the motor shortens the tether contour length. Experimental conditions influence the achievable step length resolution, and ideally experiments should be conducted with high clamp-force using slow motors linked to small beads via stiff short tethers. We focus on the limitations that the polymer-track flexibility, the thermal motion of the microsphere, and the motor kinetics pose for step-length measurement in a typical optical tweezers experiment. An expression for the signal/noise ratio in a constant-force, worm-like chain tethered particle, single-molecule experiment is developed. The signal/noise ratio is related to the Fourier transform of the pairwise distance distribution, commonly used to determine step length from a time-series. Monte Carlo simulations verify the proposed theory for experimental parameter values typically encountered with molecular motors (polymerases and helicases) translocating along single- or double-stranded nucleic acids. The predictions are consistent with recent experimental results for double-stranded DNA tethers. Our results map favorable experimental conditions for observing single motor steps on various substrates but indicate that principal resolution limits are set by thermal fluctuations.
Figures







References
-
- Yin, H., M. D. Wang, K. Svoboda, R. Landick, S. M. Block, and J. Gelles. 1995. Transcription against an applied force. Science. 270:1653–1657. - PubMed
-
- Skinner, G. M., C. G. Baumann, D. M. Quinn, J. E. Molloy, and J. G. Hoggett. 2004. Promoter binding, initiation, and elongation by bacteriophage T7 RNA polymerase. A single-molecule view of the transcription cycle. J. Biol. Chem. 279:3239–3244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

