Histidine auxotrophy in commensal and disease-causing nontypeable Haemophilus influenzae
- PMID: 17496076
- PMCID: PMC1951860
- DOI: 10.1128/JB.00146-07
Histidine auxotrophy in commensal and disease-causing nontypeable Haemophilus influenzae
Abstract
Histidine biosynthesis is one of the best studied metabolic pathways in bacteria. Although this pathway is thought to be highly conserved within and between bacterial species, a previous study identified a genetic region within the histidine operon (his) of nontypeable strains of Haemophilus influenzae (NTHI) that was more prevalent among otitis media strains than among throat commensal NTHI strains. In the present study, we further characterized this region and showed that genes in the complete his operon (hisG, -D, -C, -NB, -H, -A, -F, and -IE) are >99% conserved among four fully sequenced NTHI strains, are present in the same location in these four genomes, and are situated in the same gene order. Using PCR and dot blot hybridization, we determined that the his operon was significantly more prevalent in otitis media NTHI strains (106/121; 87.7%) than in throat strains (74/137; 54%) (prevalence ratio, 1.62; P<0.0001), suggesting a possible role in middle ear survival and/or acute otitis media. NTHI strains lacking the his operon showed attenuated growth in histidine-restricted media, confirming them as his-negative auxotrophs. Our results suggest that the ability to make histidine is an important factor in bacterial growth and survival in the middle ear, where nutrients such as histidine may be found in limited amounts. Those isolates lacking the histidine pathway were still able to survive well in the throat, which suggests that histidine is readily available in the throat environment.
Figures
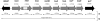
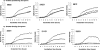
References
-
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
-
- Bayliss, C. D., D. Field, X. de Bolle, and E. R. Moxon. 2000. The generation of diversity by Haemophilus influenzae: response. Trends Microbiol. 8:435-436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

