Temperature-hypersensitive sites of the flagellar switch component FliG in Salmonella enterica serovar typhimurium
- PMID: 17496083
- PMCID: PMC1951853
- DOI: 10.1128/JB.00061-07
Temperature-hypersensitive sites of the flagellar switch component FliG in Salmonella enterica serovar typhimurium
Abstract
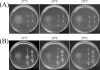
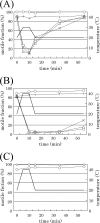
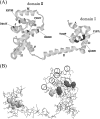
Three flagellar proteins, FliG, FliM, and FliN (FliGMN), are the components of the C ring of the flagellar motor. The genes encoding these proteins are multifunctional; they show three different phenotypes (Fla(-), Mot(-), and Che(-)), depending on the sites and types of mutations. Some of the Mot(-) mutants previously characterized are found to be motile. Reexamination of all Mot(-) mutants in fliGMN genes so far studied revealed that many of them are actually temperature sensitive (TS); that is, they are motile at 20 degrees C but nonmotile at 37 degrees C. There were two types of TS mutants: one caused a loss of function that was not reversed by a return to the permissive temperature (rigid TS), and the other caused a loss that was reversed (hyper-TS). The rigid TS mutants showed an all-or-none phenotype; that is, once a structure was formed, the structure and function were stable against temperature shifts. All of fliM and fliN and most of the fliG TS mutants belong to this group. On the other hand, the hyper-TS mutants (three of the fliG mutants) showed a temporal swimming/stop phenotype, responding to temporal temperature shifts when the structure was formed at a permissive temperature. Those hyper-TS mutation sites are localized in the C-terminal domain of the FliG molecules at sites that are different from the previously proposed functional sites. We discuss a role for this new region of FliG in the torque generation of the flagellar motor.
Figures

References
-
- Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

