Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks
- PMID: 17496093
- PMCID: PMC1951864
- DOI: 10.1128/JB.00332-07
Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks
Abstract
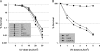
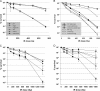
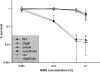
Bacterial nonhomologous end joining (NHEJ) is a recently described DNA repair pathway best characterized in mycobacteria. Bacterial NHEJ proteins LigD and Ku have been analyzed biochemically, and their roles in linear plasmid repair in vivo have been verified genetically; yet the contributions of NHEJ to repair of chromosomal DNA damage are unknown. Here we use an extensive set of NHEJ- and homologous recombination (HR)-deficient Mycobacterium smegmatis strains to probe the importance of HR and NHEJ in repairing diverse types of chromosomal DNA damage. An M. smegmatis Delta recA Delta ku double mutant has no apparent growth defect in vitro. Loss of the NHEJ components Ku and LigD had no effect on sensitivity to UV radiation, methyl methanesulfonate, or quinolone antibiotics. NHEJ deficiency had no effect on sensitivity to ionizing radiation in logarithmic- or early-stationary-phase cells but was required for ionizing radiation resistance in late stationary phase in 7H9 but not LB medium. In addition, NHEJ components were required for repair of I-SceI mediated chromosomal double-strand breaks (DSBs), and in the absence of HR, the NHEJ pathway rapidly mutates the chromosomal break site. The molecular outcomes of NHEJ-mediated chromosomal DSB repair involve predominantly single-nucleotide insertions at the break site, similar to previous findings using plasmid substrates. These findings demonstrate that prokaryotic NHEJ is specifically required for DSB repair in late stationary phase and can mediate mutagenic repair of homing endonuclease-generated chromosomal DSBs.
Figures



References
-
- Adachi, N., H. Suzuki, S. Iiizumi, and H. Koyama. 2003. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 278:35897-35902. - PubMed
-
- Boshoff, H. I., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

