The two AGPase subunits evolve at different rates in angiosperms, yet they are equally sensitive to activity-altering amino acid changes when expressed in bacteria
- PMID: 17496118
- PMCID: PMC1913735
- DOI: 10.1105/tpc.106.049676
The two AGPase subunits evolve at different rates in angiosperms, yet they are equally sensitive to activity-altering amino acid changes when expressed in bacteria
Abstract
The rate of protein evolution is generally thought to reflect, at least in part, the proportion of amino acids within the protein that are needed for proper function. In the case of ADP-glucose pyrophosphorylase (AGPase), this premise led to the hypothesis that, because the AGPase small subunit is more conserved compared with the large subunit, a higher proportion of the amino acids of the small subunit are required for enzyme activity compared with the large subunit. Evolutionary analysis indicates that the AGPase small subunit has been subject to more intense purifying selection than the large subunit in the angiosperms. However, random mutagenesis and expression of the maize (Zea mays) endosperm AGPase in bacteria show that the two AGPase subunits are equally predisposed to enzyme activity-altering amino acid changes when expressed in one environment with a single complementary subunit. As an alternative hypothesis, we suggest that the small subunit exhibits more evolutionary constraints in planta than does the large subunit because it is less tissue specific and thus must form functional enzyme complexes with different large subunits. Independent approaches provide data consistent with this alternative hypothesis.
Figures

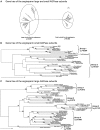




References
-
- Akashi, H. (2001). Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11 660–666. - PubMed
-
- Akihiro, T., Mizuno, K., and Fujimura, T. (2005). Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol. 46 937–946. - PubMed
-
- Bae, J.M., Giroux, M.J., and Hannah, L.C. (1990). Cloning and molecular characterization of the brittle-2 gene of maize. Maydica 35 317–322.
-
- Baldauf, S.L., Roger, A.J., Wenk-Siefert, I., and Doolittle, W.F. (2000). A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290 972–977. - PubMed
-
- Ballicora, M.A., Dubay, J.R., Devillers, C.H., and Preiss, J. (2005). Resurrecting the ancestral enzymatic role of a modulatory subunit. J. Biol. Chem. 280 10189–10195. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

