Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants
- PMID: 17496122
- PMCID: PMC1913729
- DOI: 10.1105/tpc.106.042903
Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants
Abstract
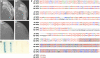
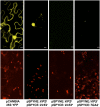
Agrobacterium tumefaciens-mediated genetic transformation is an efficient tool for genetic engineering of plants. VirE2 is a single-stranded DNA binding Agrobacterium protein that is transported into the plant cell and presumably protects the T-DNA from degradation. Using a yeast two-hybrid system, we identified Arabidopsis thaliana VIRE2-INTERACTING PROTEIN2 (VIP2) with a NOT domain that is conserved in both plants and animals. Furthermore, we provide evidence supporting VIP2 interaction with VIP1, a basic domain/leucine zipper motif-containing protein required for nuclear import and integration of T-DNA. Virus-induced gene silencing of VIP2 in Nicotiana benthamiana and characterization of the Arabidopsis vip2 mutant (At vip2) demonstrate that VIP2 is required for Agrobacterium-mediated stable transformation but not for transient transformation. Assays based upon a promoter-trap vector and quantification of T-DNA integration further confirmed VIP2 involvement in T-DNA integration. Interestingly, VIP2 transcripts were induced to a greater extent over prolonged periods after infection with a T-DNA transfer-competent Agrobacterium strain compared with the transfer-deficient Agrobacterium strain. Transcriptome analyses of At vip2 suggest that VIP2 is likely a transcriptional regulator, and the recalcitrancy to transformation in At vip2 is probably due to the combination of muted gene expression response upon Agrobacterium infection and repression of histone genes resulting in decreased T-DNA integration events.
Figures






References
-
- Anand, A., and Mysore, K.S. (2006). Agrobacterium-biology and crown gall disease. In Plant-Associated Bacteria, S.S. Gnanamanickam, ed (Dordrecht, The Netherlands: Springer Science), pp. 359–384.
-
- Anand, A., Trick, H.N., Gill, B.S., and Muthukrishnan, S. (2003. b). Stable transgene expression and random gene silencing in wheat. Plant Biotechnol. J. 1 241–252. - PubMed
-
- Anand, A., Zarir, V., Ryu, C.M., Kang, L., del-Pozo, O., Martin, G.B., and Mysore, K.S. (2007). Identification of plant genes involved in Agrobacterium-mediated transformation by using virus-induced gene silencing as a functional genomics tool. Mol. Plant Microbe Interact. 20 41–52. - PubMed
-
- Anand, A., Zhou, T., Trick, H.N., Gill, B.S., Bockus, W.W., and Muthukrishnan, S. (2003. a). Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J. Exp. Bot. 54 1101–1111. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

