Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin
- PMID: 17496129
- PMCID: PMC1932801
- DOI: 10.1128/AEM.00485-07
Identification of the biosynthetic gene cluster and an additional gene for resistance to the antituberculosis drug capreomycin
Abstract
Capreomycin (CMN) belongs to the tuberactinomycin family of nonribosomal peptide antibiotics that are essential components of the drug arsenal for the treatment of multidrug-resistant tuberculosis. Members of this antibiotic family target the ribosomes of sensitive bacteria and disrupt the function of both subunits of the ribosome. Resistance to these antibiotics in Mycobacterium species arises due to mutations in the genes coding for the 16S or 23S rRNA but can also arise due to mutations in a gene coding for an rRNA-modifying enzyme, TlyA. While Mycobacterium species develop resistance due to alterations in the drug target, it has been proposed that the CMN-producing bacterium, Saccharothrix mutabilis subsp. capreolus, uses CMN modification as a mechanism for resistance rather than ribosome modification. To better understand CMN biosynthesis and resistance in S. mutabilis subsp. capreolus, we focused on the identification of the CMN biosynthetic gene cluster in this bacterium. Here, we describe the cloning and sequence analysis of the CMN biosynthetic gene cluster from S. mutabilis subsp. capreolus ATCC 23892. We provide evidence for the heterologous production of CMN in the genetically tractable bacterium Streptomyces lividans 1326. Finally, we present data supporting the existence of an additional CMN resistance gene. Initial work suggests that this resistance gene codes for an rRNA-modifying enzyme that results in the formation of CMN-resistant ribosomes that are also resistant to the aminoglycoside antibiotic kanamycin. Thus, S. mutabilis subsp. capreolus may also use ribosome modification as a mechanism for CMN resistance.
Figures
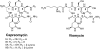


References
-
- Bibb, M. J., J. M. Ward, and S. N. Cohen. 1985. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol. Gen. Genet. 199:26-36. - PubMed
-
- Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. - PubMed
-
- Carter, J. H., II, R. H. Du Bus, J. R. Dyer, J. C. Floyd, K. C. Rice, and P. D. Shaw. 1974. Biosynthesis of viomycin. I. Origin of alpha, beta-diaminopropionic acid and serine. Biochemistry 13:1221-1227. - PubMed
-
- Carter, J. H., II, R. H. Du Bus, J. R. Dyer, J. C. Floyd, K. C. Rice, and P. D. Shaw. 1974. Biosynthesis of viomycin. II. Origin of beta-lysine and viomycidine. Biochemistry 13:1227-1233. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

