Protein arginine deiminase 4: evidence for a reverse protonation mechanism
- PMID: 17497940
- PMCID: PMC2212595
- DOI: 10.1021/bi700095s
Protein arginine deiminase 4: evidence for a reverse protonation mechanism
Abstract
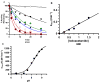
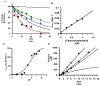
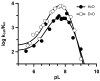
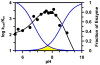
The presumed role of an overactive protein arginine deiminase 4 (PAD4) in the pathophysiology of rheumatoid arthritis (RA) suggests that PAD4 inhibitors could be used to treat an underlying cause of RA, potentially offering a mechanism to stop further disease progression. Thus, the development of such inhibitors is of paramount importance. Toward the goal of developing such inhibitors, we initiated efforts to characterize the catalytic mechanism of PAD4 and thereby identify important mechanistic features that can be exploited for inhibitor development. Herein we report the results of mutagenesis studies as well as our efforts to characterize the initial steps of the PAD4 reaction, in particular, the protonation status of Cys645 and His471 prior to substrate binding. The results indicate that Cys645, the active site nucleophile, exists as the thiolate in the active form of the free enzyme. pH studies on PAD4 further suggest that this enzyme utilizes a reverse protonation mechanism.
Figures
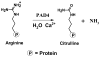

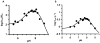

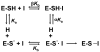
References
-
- Finesilver AG. Newer approaches to the treatment of rheumatoid arthritis. Wisconsin Medical Journal. 2003;102:34–37. - PubMed
-
- Smolen JS, Steiner G. Therapeutic strategies for rheumatoid arthritis. Nat Rev Drug Discov. 2003;2:473–488. - PubMed
-
- Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono M, Ohtsuki M, Furukawa H, Yoshino S, Yukioka M, Tohma S, Matsubara T, Wakitani S, Teshima R, Nishioka Y, Sekine A, Iida A, Takahashi A, Tsunoda T, Nakamura Y, Yamamoto K. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. - PubMed
-
- Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–1118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

