Species-wide distribution of highly polymorphic minisatellite markers suggests past and present genetic exchanges among house mouse subspecies
- PMID: 17501990
- PMCID: PMC1929145
- DOI: 10.1186/gb-2007-8-5-r80
Species-wide distribution of highly polymorphic minisatellite markers suggests past and present genetic exchanges among house mouse subspecies
Abstract
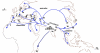
Background: Four hypervariable minisatellite loci were scored on a panel of 116 individuals of various geographical origins representing a large part of the diversity present in house mouse subspecies. Internal structures of alleles were determined by minisatellite variant repeat mapping PCR to produce maps of intermingled patterns of variant repeats along the repeat array. To reconstruct the genealogy of these arrays of variable length, the specifically designed software MS_Align was used to estimate molecular divergences, graphically represented as neighbor-joining trees.
Results: Given the high haplotypic diversity detected (mean He = 0.962), these minisatellite trees proved to be highly informative for tracing past and present genetic exchanges. Examples of identical or nearly identical alleles were found across subspecies and in geographically very distant locations, together with poor lineage sorting among subspecies except for the X-chromosome locus MMS30 in Mus mus musculus. Given the high mutation rate of mouse minisatellite loci, this picture cannot be interpreted only with simple splitting events followed by retention of polymorphism, but implies recurrent gene flow between already differentiated entities.
Conclusion: This strongly suggests that, at least for the chromosomal regions under scrutiny, wild house mouse subspecies constitute a set of interrelated gene pools still connected through long range gene flow or genetic exchanges occurring in the various contact zones existing nowadays or that have existed in the past. Identifying genomic regions that do not follow this pattern will be a challenging task for pinpointing genes important for speciation.
Figures







References
-
- Tamaki K, Jeffreys AJ. Human tandem repeat sequences in forensic DNA typing. Leg Med (Tokyo) 2005;7:244–250. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

