Promoter and transcription analysis of penicillin-binding protein genes in Streptococcus gordonii
- PMID: 17502405
- PMCID: PMC1932516
- DOI: 10.1128/AAC.01127-06
Promoter and transcription analysis of penicillin-binding protein genes in Streptococcus gordonii
Abstract
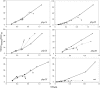
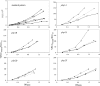
An optimally cross-linked peptidoglycan requires both transglycosylation and transpeptidation, provided by class A and class B penicillin-binding proteins (PBPs). Streptococcus gordonii possesses three class A PBPs (PBPs 1A, 1B, and 2A) and two class B PBPs (PBPs 2B and 2X) that are important for penicillin resistance. High-level resistance (MIC, > or =2 microg/ml) requires mutations in class B PBPs. However, although unmutated, class A PBPs are critical to facilitate resistance development (M. Haenni and P. Moreillon, Antimicrob. Agents Chemother. 50:4053-4061, 2006). Thus, their overexpression might be important to sustain the drug. Here, we determined the promoter regions of the S. gordonii PBPs and compared them to those of other streptococci. The extended -10 box was highly conserved and complied with a sigma(A)-type promoter consensus sequence. In contrast, the -35 box was poorly conserved, leaving the possibility of differential PBP regulation. Gene expression in a penicillin-susceptible parent (MIC, 0.008 microg/ml) and a high-level-resistant mutant (MIC, 2 microg/ml) was monitored using luciferase fusions. In the absence of penicillin, all PBPs were constitutively expressed, but their expression was globally increased (1.5 to 2 times) in the resistant mutant. In the presence of penicillin, class A PBPs were specifically overexpressed both in the parent (PBP 2A) and in the resistant mutant (PBPs 1A and 2A). By increasing transglycosylation, class A PBPs could promote peptidoglycan stability when transpeptidase is inhibited by penicillin. Since penicillin-related induction of class A PBPs occurred in both susceptible and resistant cells, such a mutation-independent facilitating mechanism could be operative at each step of resistance development.
Figures




Similar articles
-
Highly variable penicillin resistance determinants PBP 2x, PBP 2b, and PBP 1a in isolates of two Streptococcus pneumoniae clonal groups, Poland 23F-16 and Poland 6B-20.Antimicrob Agents Chemother. 2008 Mar;52(3):1021-7. doi: 10.1128/AAC.01082-07. Epub 2007 Dec 26. Antimicrob Agents Chemother. 2008. PMID: 18160523 Free PMC article.
-
Mutations in penicillin-binding protein (PBP) genes and in non-PBP genes during selection of penicillin-resistant Streptococcus gordonii.Antimicrob Agents Chemother. 2006 Dec;50(12):4053-61. doi: 10.1128/AAC.00676-06. Epub 2006 Sep 25. Antimicrob Agents Chemother. 2006. PMID: 17000741 Free PMC article.
-
Combinations of PBPs and MurM protein variants in early and contemporary high-level penicillin-resistant Streptococcus pneumoniae isolates in Spain.J Antimicrob Chemother. 2006 May;57(5):983-6. doi: 10.1093/jac/dkl083. Epub 2006 Mar 13. J Antimicrob Chemother. 2006. PMID: 16533824
-
Penicillin-binding proteins and beta-lactam resistance.FEMS Microbiol Rev. 2008 Mar;32(2):361-85. doi: 10.1111/j.1574-6976.2007.00095.x. Epub 2008 Jan 29. FEMS Microbiol Rev. 2008. PMID: 18248419 Review.
-
Methicillin-resistant Staphylococcus aureus. Mechanisms of resistance and implications for treatment.Postgrad Med. 2001 Feb;109(2 Suppl):43-50. doi: 10.3810/pgm.02.2001.suppl12.65. Postgrad Med. 2001. PMID: 19667557 Review.
Cited by
-
A cryptic oxidoreductase safeguards oxidative protein folding in Corynebacterium diphtheriae.Proc Natl Acad Sci U S A. 2023 Feb 21;120(8):e2208675120. doi: 10.1073/pnas.2208675120. Epub 2023 Feb 14. Proc Natl Acad Sci U S A. 2023. PMID: 36787356 Free PMC article.
-
Fitness cost and impaired survival in penicillin-resistant Streptococcus gordonii isolates selected in the laboratory.Antimicrob Agents Chemother. 2008 Jan;52(1):337-9. doi: 10.1128/AAC.00939-07. Epub 2007 Nov 12. Antimicrob Agents Chemother. 2008. PMID: 17999969 Free PMC article.
-
Genetic Loci Associated With Fluoride Resistance in Streptococcus mutans.Front Microbiol. 2018 Dec 11;9:3093. doi: 10.3389/fmicb.2018.03093. eCollection 2018. Front Microbiol. 2018. PMID: 30619172 Free PMC article.
-
Promoter Identification and Transcription Analysis of Penicillin-Binding Protein Genes in Streptococcus pneumoniae R6.Microb Drug Resist. 2016 Sep;22(6):487-98. doi: 10.1089/mdr.2016.0084. Epub 2016 Jul 13. Microb Drug Resist. 2016. PMID: 27409661 Free PMC article.
-
A low-affinity penicillin-binding protein 2x variant is required for heteroresistance in Streptococcus pneumoniae.Antimicrob Agents Chemother. 2014 Jul;58(7):3934-41. doi: 10.1128/AAC.02547-14. Epub 2014 Apr 28. Antimicrob Agents Chemother. 2014. PMID: 24777105 Free PMC article.
References
-
- Bizzini, A., P. Majcherczyk, S. Beggah-Moller, B. Soldo, J. M. Entenza, M. Gaillard, P. Moreillon, and V. Lazarevic. 2007. Effects of alpha-phosphoglucomutase deficiency on cell wall properties and fitness in Streptococcus gordonii. Microbiology 153:490-498. - PubMed
-
- Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45:1267-1276. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

