Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins
- PMID: 17502600
- PMCID: PMC1885576
- DOI: 10.1073/pnas.0701453104
Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins
Abstract
Peptidoglycan recognition proteins (PGRPs) are highly conserved pattern-recognition molecules of the innate immune system that bind bacterial peptidoglycans (PGNs), which are polymers of alternating N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) cross-linked by short peptide stems. Human PRGPs are bactericidal against pathogenic and nonpathogenic Gram-positive bacteria, but not normal flora bacteria. Like certain glycopeptide antibiotics (e.g., vancomycin), PGRPs kill bacteria by directly interacting with their cell wall PGN, thereby interfering with PGN maturation. To better understand the bactericidal mechanism of PGRPs, we determined the crystal structure of the C-terminal PGN-binding domain of human PGRP-I beta in complex with NAG-NAM-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala, a synthetic glycopeptide comprising a complete PGN repeat. This structure, in conjunction with the previously reported NMR structure of a dimeric PGN fragment, permitted identification of major conformational differences between free and PGRP-bound PGN with respect to the relative orientation of saccharide and peptide moieties. These differences provided structural insights into the bactericidal mechanism of human PGRPs. On the basis of molecular modeling, we propose that these proteins disrupt cell wall maturation not only by sterically encumbering access of biosynthetic enzymes to the nascent PGN chains, but also by locking PGN into a conformation that prevents formation of cross-links between peptide stems in the growing cell wall.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

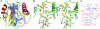
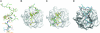
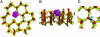
Similar articles
-
Crystal structure of human peptidoglycan recognition protein I alpha bound to a muramyl pentapeptide from Gram-positive bacteria.Protein Sci. 2006 May;15(5):1199-206. doi: 10.1110/ps.062077606. Protein Sci. 2006. PMID: 16641493 Free PMC article.
-
Structural basis for peptidoglycan binding by peptidoglycan recognition proteins.Proc Natl Acad Sci U S A. 2004 Dec 7;101(49):17168-73. doi: 10.1073/pnas.0407856101. Epub 2004 Nov 30. Proc Natl Acad Sci U S A. 2004. PMID: 15572450 Free PMC article.
-
Crystal structure of human peptidoglycan recognition protein S (PGRP-S) at 1.70 A resolution.J Mol Biol. 2005 Apr 8;347(4):683-91. doi: 10.1016/j.jmb.2005.01.070. J Mol Biol. 2005. PMID: 15769462
-
Peptidoglycan recognition proteins of the innate immune system.Trends Microbiol. 2007 Mar;15(3):127-34. doi: 10.1016/j.tim.2007.01.006. Epub 2007 Feb 1. Trends Microbiol. 2007. PMID: 17275309 Review.
-
Peptidoglycan recognition proteins in Drosophila immunity.Dev Comp Immunol. 2014 Jan;42(1):36-41. doi: 10.1016/j.dci.2013.06.006. Epub 2013 Jun 22. Dev Comp Immunol. 2014. PMID: 23796791 Free PMC article. Review.
Cited by
-
Key side products due to reactivity of dimethylmaleoyl moiety as amine protective group.Chem Zvesti. 2009 Oct;63(5):592-597. doi: 10.2478/s11696-009-0048-0. Epub 2009 Aug 25. Chem Zvesti. 2009. PMID: 31920214 Free PMC article.
-
Bacterial peptidoglycan degrading enzymes and their impact on host muropeptide detection.J Innate Immun. 2009;1(2):88-97. doi: 10.1159/000181181. J Innate Immun. 2009. PMID: 19319201 Free PMC article. Review.
-
Success stories of natural product-derived compounds from plants as multidrug resistance modulators in microorganisms.RSC Adv. 2023 Mar 8;13(12):7798-7817. doi: 10.1039/d3ra00184a. eCollection 2023 Mar 8. RSC Adv. 2023. PMID: 36909750 Free PMC article. Review.
-
Crystal structures of penicillin-binding protein 6 from Escherichia coli.J Am Chem Soc. 2009 Oct 14;131(40):14345-54. doi: 10.1021/ja903773f. J Am Chem Soc. 2009. PMID: 19807181 Free PMC article.
-
Self-assembled and Zn(II)-coordinated dipeptide nanoparticles with membrane-rupturing action on bacteria.Appl Microbiol Biotechnol. 2023 Sep;107(18):5775-5787. doi: 10.1007/s00253-023-12648-4. Epub 2023 Jul 13. Appl Microbiol Biotechnol. 2023. PMID: 37439833
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

