The frequency and structure of recombinant products is determined by the cellular level of MutL
- PMID: 17502621
- PMCID: PMC1885606
- DOI: 10.1073/pnas.0610149104
The frequency and structure of recombinant products is determined by the cellular level of MutL
Abstract
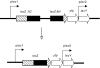
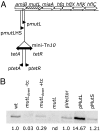
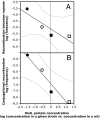
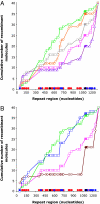
The presence of repeated DNA sequences is a genomic liability, because interrepeat recombination can result in chromosomal rearrangements. The mismatch repair system prevents recombination between nonidentical repeats, but the mechanism of antirecombination has not been established. Although the MutS protein binds to base pair mismatches in heteroduplex DNA, the role of the MutL protein in preventing recombination is unknown. In a screen designed to identify new cellular functions that suppress deletion formation involving nonidentical DNA repeats, we isolated a mutL mutant having a separation-of-function phenotype. The mutant showed an increased frequency of deletions but not of mutations. The split phenotype is due to a decreased MutL level, indicating that recombination, but not replication editing, is highly sensitive to MutL level. By altering the MutL level, we found that the frequency of deletion-generating recombination is inversely related to the amount of cellular MutL. DNA sequence analysis of the recombined repeats shows that the tolerance of base pair mismatches in heteroduplex DNA is also inversely correlated with MutL level. Unlike recombination, correction of misincorporation errors by mismatch repair is insensitive to fluctuations in MutL level. Overproduction of MutS does not affect either of these phenotypes, suggesting that, unlike MutL, MutS is not limiting for mismatch repair activities. These results indicate that MutL (i) determines effective DNA homology in recombination processes and (ii) fine tunes the process of deletion formation involving repeated, diverged DNA sequences.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Dominant negative mutator mutations in the mutS gene of Escherichia coli.J Bacteriol. 1994 Sep;176(17):5393-400. doi: 10.1128/jb.176.17.5393-5400.1994. J Bacteriol. 1994. PMID: 8071216 Free PMC article.
-
In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination.DNA Repair (Amst). 2003 Apr 2;2(4):387-405. doi: 10.1016/s1568-7864(02)00245-8. DNA Repair (Amst). 2003. PMID: 12606120
-
Characterization of the defects in the ATP lid of E. coli MutL that cause transient hypermutability.DNA Repair (Amst). 2013 Oct;12(10):864-9. doi: 10.1016/j.dnarep.2013.07.003. Epub 2013 Aug 2. DNA Repair (Amst). 2013. PMID: 23916559
-
Wot the 'L-Does MutL do?Mutat Res. 2010 Dec;705(3):228-38. doi: 10.1016/j.mrrev.2010.07.002. Epub 2010 Aug 3. Mutat Res. 2010. PMID: 20667509 Review.
-
MutL: conducting the cell's response to mismatched and misaligned DNA.Bioessays. 2010 Jan;32(1):51-9. doi: 10.1002/bies.200900089. Bioessays. 2010. PMID: 19953589 Review.
Cited by
-
Gene Expression Noise Produces Cell-to-Cell Heterogeneity in Eukaryotic Homologous Recombination Rate.Front Genet. 2019 May 21;10:475. doi: 10.3389/fgene.2019.00475. eCollection 2019. Front Genet. 2019. PMID: 31164905 Free PMC article.
-
Involvement of mismatch repair in the reciprocal control of motility and adherence of uropathogenic Escherichia coli.Infect Immun. 2012 Jun;80(6):1969-79. doi: 10.1128/IAI.00043-12. Epub 2012 Apr 2. Infect Immun. 2012. PMID: 22473602 Free PMC article.
-
Mismatch repair inhibits homeologous recombination via coordinated directional unwinding of trapped DNA structures.Mol Cell. 2013 Aug 8;51(3):326-37. doi: 10.1016/j.molcel.2013.07.008. Mol Cell. 2013. PMID: 23932715 Free PMC article.
-
Escherichia coli YafP protein modulates DNA damaging property of the nitroaromatic compounds.Nucleic Acids Res. 2011 May;39(10):4192-201. doi: 10.1093/nar/gkr050. Epub 2011 Feb 7. Nucleic Acids Res. 2011. PMID: 21300638 Free PMC article.
-
Effect of subinhibitory concentrations of antibiotics on intrachromosomal homologous recombination in Escherichia coli.Antimicrob Agents Chemother. 2009 Aug;53(8):3411-5. doi: 10.1128/AAC.00358-09. Epub 2009 Jun 1. Antimicrob Agents Chemother. 2009. PMID: 19487441 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

