A genomewide admixture map for Latino populations
- PMID: 17503322
- PMCID: PMC1867092
- DOI: 10.1086/518313
A genomewide admixture map for Latino populations
Abstract
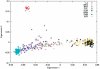
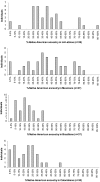
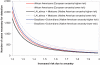
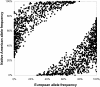
Admixture mapping is an economical and powerful approach for localizing disease genes in populations of recently mixed ancestry and has proven successful in African Americans. The method holds equal promise for Latinos, who typically inherit a mix of European, Native American, and African ancestry. However, admixture mapping in Latinos has not been practical because of the lack of a map of ancestry-informative markers validated in Native American and other populations. To address this, we screened multiple databases, containing millions of markers, to identify 4,186 markers that were putatively informative for determining the ancestry of chromosomal segments in Latino populations. We experimentally validated each of these markers in at least 232 new Latino, European, Native American, and African samples, and we selected a subset of 1,649 markers to form an admixture map. An advantage of our strategy is that we focused our map on markers distinguishing Native American from other ancestries and restricted it to markers with very similar frequencies in Europeans and Africans, which decreased the number of markers needed and minimized the possibility of false disease associations. We evaluated the effectiveness of our map for localizing disease genes in four Latino populations from both North and South America.
Figures
References
Web Resources
-
- D.R. Lab Web site, http://genepath.med.harvard.edu/~reich/Software.htm (for ANCESTRYMAP and EIGENSOFT software)
-
- Latino admixture map, http://genepath.med.harvard.edu/~reich/Latinomap.htm (for the list of 1,649 markers)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for prostate cancer, type 2 diabetes, obesity, gallbladder disease, rheumatoid arthritis, and asthma)
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

