TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer
- PMID: 17504517
- PMCID: PMC1929092
- DOI: 10.1186/bcr1675
TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer
Abstract
Introduction: Gene expression profiling of breast carcinomas has increased our understanding of the heterogeneous biology of this disease and promises to impact clinical care. The aim of this study was to evaluate the prognostic value of gene expression-based classification along with established prognostic markers and mutation status of the TP53 gene (tumour protein p53) in a group of breast cancer patients with long-term (12 to 16 years) follow-up.
Methods: The clinical and histopathological parameters of 200 breast cancer patients were studied for their effects on clinical outcome using univariate/multivariate Cox regression. The prognostic impact of mutations in the TP53 gene, identified using temporal temperature gradient gel electrophoresis and sequencing, was also evaluated. Eighty of the samples were analyzed for gene expression using 42 K cDNA microarrays and the patients were assigned to five previously defined molecular expression groups. The strength of the gene expression based classification versus standard markers was evaluated by adding this variable to the Cox regression model used to analyze all samples.
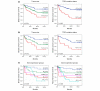
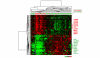
Results: Both univariate and multivariate analysis showed that TP53 mutation status, tumor size and lymph node status were the strongest predictors of breast cancer survival for the whole group of patients. Analyses of the patients with gene expression data showed that TP53 mutation status, gene expression based classification, tumor size and lymph node status were significant predictors of survival. Breast cancer cases in the 'basal-like' and 'ERBB2+' gene expression subgroups had a very high mortality the first two years, while the 'highly proliferating luminal' cases developed the disease more slowly, showing highest mortality after 5 to 8 years. The TP53 mutation status showed strong association with the 'basal-like' and 'ERBB2+' subgroups, and tumors with mutation had a characteristic gene expression pattern.
Conclusion: TP53 mutation status and gene-expression based groups are important survival markers of breast cancer, and these molecular markers may provide prognostic information that complements clinical variables. The study adds experience and knowledge to an ongoing characterization and classification of the disease.
Figures



Similar articles
-
Combined Immunohistochemistry of PLK1, p21, and p53 for Predicting TP53 Status: An Independent Prognostic Factor of Breast Cancer.Am J Surg Pathol. 2015 Aug;39(8):1026-34. doi: 10.1097/PAS.0000000000000386. Am J Surg Pathol. 2015. PMID: 26171916
-
TP53 mutation is an independent prognostic marker for poor outcome in both node-negative and node-positive breast cancer.Acta Oncol. 2000;39(3):327-33. doi: 10.1080/028418600750013096. Acta Oncol. 2000. PMID: 10987229
-
Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen.PLoS Med. 2007 Mar;4(3):e90. doi: 10.1371/journal.pmed.0040090. PLoS Med. 2007. PMID: 17388661 Free PMC article.
-
p53 in breast cancer subtypes and new insights into response to chemotherapy.Breast. 2013 Aug;22 Suppl 2:S27-9. doi: 10.1016/j.breast.2013.07.005. Breast. 2013. PMID: 24074787 Review.
-
Overview on human breast cancer with focus on prognostic and predictive factors with special attention on the tumour suppressor gene p53.Acta Oncol. 1996;35 Suppl 5:96-102. doi: 10.3109/02841869609083980. Acta Oncol. 1996. PMID: 9142977 Review.
Cited by
-
Prevalence of TP53 germ line mutations in young Pakistani breast cancer patients.Fam Cancer. 2012 Jun;11(2):307-11. doi: 10.1007/s10689-012-9509-7. Fam Cancer. 2012. PMID: 22311583
-
BST-2/tetherin is overexpressed in mammary gland and tumor tissues in MMTV-induced mammary cancer.Virology. 2013 Sep;444(1-2):124-39. doi: 10.1016/j.virol.2013.05.042. Epub 2013 Jun 25. Virology. 2013. PMID: 23806386 Free PMC article.
-
Integrative functional genomics analysis of sustained polyploidy phenotypes in breast cancer cells identifies an oncogenic profile for GINS2.Neoplasia. 2010 Nov;12(11):877-88. doi: 10.1593/neo.10548. Neoplasia. 2010. PMID: 21082043 Free PMC article.
-
Insight into the heterogeneity of breast cancer through next-generation sequencing.J Clin Invest. 2011 Oct;121(10):3810-8. doi: 10.1172/JCI57088. Epub 2011 Oct 3. J Clin Invest. 2011. PMID: 21965338 Free PMC article. Review.
-
Circadian PERformance in breast cancer: a germline and somatic genetic study of PER3VNTR polymorphisms and gene co-expression.NPJ Breast Cancer. 2021 Sep 10;7(1):118. doi: 10.1038/s41523-021-00329-2. NPJ Breast Cancer. 2021. PMID: 34508103 Free PMC article.
References
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de RM, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

