Sodium-dependent extracellular accessibility of Lys-84 in the sodium/dicarboxylate cotransporter
- PMID: 17504760
- PMCID: PMC2864014
- DOI: 10.1074/jbc.M701113200
Sodium-dependent extracellular accessibility of Lys-84 in the sodium/dicarboxylate cotransporter
Abstract
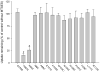
The Na(+)/dicarboxylate cotransporter transports Na(+) with citric acid cycle intermediates such as succinate and citrate. The present study focuses on transmembrane helix 3, which is highly conserved among the members of the SLC13 family. Fifteen amino acids in the extracellular half of transmembrane helix (amino acids 98-112) as well as Lys-84, previously shown to affect substrate affinity, were mutated individually to cysteine and expressed in the human retinal pigment epithelial cell line. Transport specificity ratio analysis shows that determinants for distinguishing succinate and citrate are found at amino acids Lys-84, Glu-101, Trp-103, His-106, and Leu-111. All of the mutants were tested for sensitivity to the membrane-impermeant cysteine-specific reagent (2-sulfonatoethyl) methanethiosulfonate (MTSES), but only K84C was sensitive to MTSES inhibition. The sensitivity of K84C to MTSES was greatest in the presence of sodium, and the inhibition could be prevented by addition of substrate or replacement of sodium, indicating that the accessibility of Lys-84 changes with conformational state. The substrate protection of MTSES inhibition of K84C appears to occur early in the transport cycle, before the large-scale conformational change associated with translocation of substrate. The results point to a new location for Lys-84 within the substrate access pore of the Na(+)/dicarboxylate cotransporter, either in a transmembrane helix or a reentrant loop facing a water-filled pore.
Figures
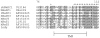


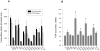

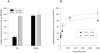


Similar articles
-
Transmembrane helix 7 in the Na+/dicarboxylate cotransporter 1 is an outer helix that contains residues critical for function.Biochim Biophys Acta. 2011 Jun;1808(6):1454-61. doi: 10.1016/j.bbamem.2010.11.007. Epub 2010 Nov 10. Biochim Biophys Acta. 2011. PMID: 21073858 Free PMC article.
-
Sodium-coupled dicarboxylate and citrate transporters from the SLC13 family.Pflugers Arch. 2014 Jan;466(1):119-30. doi: 10.1007/s00424-013-1369-y. Epub 2013 Oct 10. Pflugers Arch. 2014. PMID: 24114175 Review.
-
Conformationally sensitive residues in extracellular loop 5 of the Na+/dicarboxylate co-transporter.J Biol Chem. 2005 May 13;280(19):18728-35. doi: 10.1074/jbc.M501265200. Epub 2005 Mar 17. J Biol Chem. 2005. PMID: 15774465 Free PMC article.
-
Determinants of substrate and cation transport in the human Na+/dicarboxylate cotransporter NaDC3.J Biol Chem. 2014 Jun 13;289(24):16998-7008. doi: 10.1074/jbc.M114.554790. Epub 2014 May 7. J Biol Chem. 2014. PMID: 24808185 Free PMC article.
-
Sodium-coupled transporters for Krebs cycle intermediates.Annu Rev Physiol. 1999;61:663-82. doi: 10.1146/annurev.physiol.61.1.663. Annu Rev Physiol. 1999. PMID: 10099705 Review.
Cited by
-
Mutations in the Na(+)/citrate cotransporter NaCT (SLC13A5) in pediatric patients with epilepsy and developmental delay.Mol Med. 2016 May 26;22:310-21. doi: 10.2119/molmed.2016.00077. Mol Med. 2016. PMID: 27261973 Free PMC article.
-
Threonine-509 is a determinant of apparent affinity for both substrate and cations in the human Na+/dicarboxylate cotransporter.Biochemistry. 2008 Jan 22;47(3):1087-93. doi: 10.1021/bi701417h. Epub 2007 Dec 28. Biochemistry. 2008. PMID: 18161988 Free PMC article.
-
Identification of conformationally sensitive amino acids in the Na(+)/dicarboxylate symporter (SdcS).Biochemistry. 2009 Apr 7;48(13):3017-24. doi: 10.1021/bi8022625. Biochemistry. 2009. PMID: 19260674 Free PMC article.
-
Transmembrane helix 7 in the Na+/dicarboxylate cotransporter 1 is an outer helix that contains residues critical for function.Biochim Biophys Acta. 2011 Jun;1808(6):1454-61. doi: 10.1016/j.bbamem.2010.11.007. Epub 2010 Nov 10. Biochim Biophys Acta. 2011. PMID: 21073858 Free PMC article.
-
Sodium-coupled dicarboxylate and citrate transporters from the SLC13 family.Pflugers Arch. 2014 Jan;466(1):119-30. doi: 10.1007/s00424-013-1369-y. Epub 2013 Oct 10. Pflugers Arch. 2014. PMID: 24114175 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

