Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo
- PMID: 17504939
- PMCID: PMC1865493
- DOI: 10.1101/gad.1528307
Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo
Abstract
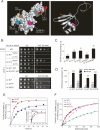
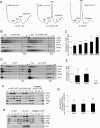
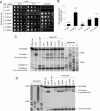
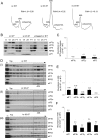
Selection of the AUG start codon is a key step in translation initiation requiring hydrolysis of GTP in the eIF2*GTP*Met-tRNA(i)(Met) ternary complex (TC) and subsequent P(i) release from eIF2*GDP*P(i). It is thought that eIF1 prevents recognition of non-AUGs by promoting scanning and blocking P(i) release at non-AUG codons. We show that Sui(-) mutations in Saccharomyces cerevisiae eIF1, which increase initiation at UUG codons, reduce interaction of eIF1 with 40S subunits in vitro and in vivo, and both defects are diminished in cells by overexpressing the mutant proteins. Remarkably, Sui(-) mutation ISQLG(93-97)ASQAA (abbreviated 93-97) accelerates eIF1 dissociation and P(i) release from reconstituted preinitiation complexes (PICs), whereas a hyperaccuracy mutation in eIF1A (that suppresses Sui(-) mutations) decreases the eIF1 off-rate. These findings demonstrate that eIF1 dissociation is a critical step in start codon selection, which is modulated by eIF1A. We also describe Gcd(-) mutations in eIF1 that impair TC loading on 40S subunits or destabilize the multifactor complex containing eIF1, eIF3, eIF5, and TC, showing that eIF1 promotes PIC assembly in vivo beyond its important functions in AUG selection.
Figures
Comment in
-
Translation factor control of ribosome conformation during start codon selection.Genes Dev. 2007 Jun 1;21(11):1280-7. doi: 10.1101/gad.1562707. Genes Dev. 2007. PMID: 17545463 No abstract available.
Similar articles
-
Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection.Mol Cell Biol. 2004 Nov;24(21):9437-55. doi: 10.1128/MCB.24.21.9437-9455.2004. Mol Cell Biol. 2004. PMID: 15485912 Free PMC article.
-
Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy.RNA. 2014 Feb;20(2):150-67. doi: 10.1261/rna.042069.113. Epub 2013 Dec 13. RNA. 2014. PMID: 24335188 Free PMC article.
-
β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40 S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo.J Biol Chem. 2013 Sep 20;288(38):27546-27562. doi: 10.1074/jbc.M113.498642. Epub 2013 Jul 26. J Biol Chem. 2013. PMID: 23893413 Free PMC article.
-
The scanning mechanism of eukaryotic translation initiation.Annu Rev Biochem. 2014;83:779-812. doi: 10.1146/annurev-biochem-060713-035802. Epub 2014 Jan 29. Annu Rev Biochem. 2014. PMID: 24499181 Review.
-
Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation.Trends Biochem Sci. 2017 Aug;42(8):589-611. doi: 10.1016/j.tibs.2017.03.004. Epub 2017 Apr 22. Trends Biochem Sci. 2017. PMID: 28442192 Review.
Cited by
-
Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode.Mol Cell Biol. 2012 Oct;32(19):3978-89. doi: 10.1128/MCB.00376-12. Epub 2012 Jul 30. Mol Cell Biol. 2012. PMID: 22851688 Free PMC article.
-
eIF1 and eIF5 dynamically control translation start site fidelity.bioRxiv [Preprint]. 2024 Jul 13:2024.07.10.602410. doi: 10.1101/2024.07.10.602410. bioRxiv. 2024. Update in: Nat Struct Mol Biol. 2025 Jul 28. doi: 10.1038/s41594-025-01629-y. PMID: 39026837 Free PMC article. Updated. Preprint.
-
Human eIF5 and eIF1A Compete for Binding to eIF5B.Biochemistry. 2018 Oct 9;57(40):5910-5920. doi: 10.1021/acs.biochem.8b00839. Epub 2018 Sep 26. Biochemistry. 2018. PMID: 30211544 Free PMC article.
-
Molecular mechanism of scanning and start codon selection in eukaryotes.Microbiol Mol Biol Rev. 2011 Sep;75(3):434-67, first page of table of contents. doi: 10.1128/MMBR.00008-11. Microbiol Mol Biol Rev. 2011. PMID: 21885680 Free PMC article. Review.
-
Toward a Kinetic Understanding of Eukaryotic Translation.Cold Spring Harb Perspect Biol. 2019 Feb 1;11(2):a032706. doi: 10.1101/cshperspect.a032706. Cold Spring Harb Perspect Biol. 2019. PMID: 29959192 Free PMC article. Review.
References
-
- Algire M.A., Maag D., Savio P., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Maag D., Savio P., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Savio P., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Acker M.G., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Tarun S.Z., Jr., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Sachs A.B., Asano K., Nielsen K.H., Olsen D.S., Phan L., Asano K., Nielsen K.H., Olsen D.S., Phan L., Nielsen K.H., Olsen D.S., Phan L., Olsen D.S., Phan L., Phan L., et al. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. - PMC - PubMed
-
- Algire M.A., Maag D., Lorsch J.R., Maag D., Lorsch J.R., Lorsch J.R. π release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. - PubMed
-
- Asano K., Phan L., Anderson J., Hinnebusch A.G., Phan L., Anderson J., Hinnebusch A.G., Anderson J., Hinnebusch A.G., Hinnebusch A.G. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:18573–18585. - PubMed
-
- Asano K., Clayton J., Shalev A., Hinnebusch A.G., Clayton J., Shalev A., Hinnebusch A.G., Shalev A., Hinnebusch A.G., Hinnebusch A.G. A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5, and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes & Dev. 2000;14:2534–2546. - PMC - PubMed
-
- Asano K., Shalev A., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Shalev A., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Phan L., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Nielsen K., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Clayton J., Valášek L., Donahue T.F., Hinnebusch A.G., Valášek L., Donahue T.F., Hinnebusch A.G., Donahue T.F., Hinnebusch A.G., Hinnebusch A.G. Multiple roles for the carboxyl terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
