Synapses: sites of cell recognition, adhesion, and functional specification
- PMID: 17506641
- PMCID: PMC3368613
- DOI: 10.1146/annurev.biochem.75.103004.142811
Synapses: sites of cell recognition, adhesion, and functional specification
Abstract
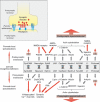
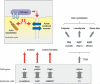
Synapses are specialized adhesive contacts characteristic of many types of cell-cell interactions involving neurons, immune cells, epithelial cells, and even pathogens and host cells. Cell-cell adhesion is mediated by structurally diverse classes of cell-surface glycoproteins, which form homophilic or heterophilic interactions across the intercellular space. Adhesion proteins bind to a cytoplasmic network of scaffolding proteins, regulators of the actin cytoskeleton, and signal transduction pathways that control the structural and functional organization of synapses. The themes of this review are to compare the organization of synapses in different cell types and to understand how different classes of cell adhesion proteins and cytoplasmic protein networks specify the assembly of functionally distinct synapses in different cell contexts.
Figures



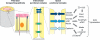
References
-
- Bennett MR. Brain Res. Bull. 1999;50:95–118. - PubMed
-
- Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Science. 1987;236:799–806. - PubMed
-
- Bruses JL, Rutishauser U. Biochimie. 2001;83:635–43. - PubMed
-
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, et al. Nature. 1994;367:455–59. - PubMed
-
- Hu H, Tomasiewicz H, Magnuson T, Rutishauser U. Neuron. 1996;16:735–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

