Regulatory conservation of protein coding and microRNA genes in vertebrates: lessons from the opossum genome
- PMID: 17506886
- PMCID: PMC1929153
- DOI: 10.1186/gb-2007-8-5-r84
Regulatory conservation of protein coding and microRNA genes in vertebrates: lessons from the opossum genome
Abstract
Background: Being the first noneutherian mammal sequenced, Monodelphis domestica (opossum) offers great potential for enhancing our understanding of the evolutionary processes that take place in mammals. This study focuses on the evolutionary relationships between conservation of noncoding sequences, cis-regulatory elements, and biologic functions of regulated genes in opossum and eight vertebrate species.
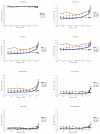
Results: Analysis of 145 intergenic microRNA and all protein coding genes revealed that the upstream sequences of the former are up to twice as conserved as the latter among mammals, except in the first 500 base pairs, where the conservation is similar. Comparison of promoter conservation in 513 protein coding genes and related transcription factor binding sites (TFBSs) showed that 41% of the known human TFBSs are located in the 6.7% of promoter regions that are conserved between human and opossum. Some core biologic processes exhibited significantly fewer conserved TFBSs in human-opossum comparisons, suggesting greater functional divergence. A new measure of efficiency in multigenome phylogenetic footprinting (base regulatory potential rate [BRPR]) shows that including human-opossum conservation increases specificity in finding human TFBSs.
Conclusion: Opossum facilitates better estimation of promoter conservation and TFBS turnover among mammals. The fact that substantial TFBS numbers are located in a small proportion of the human-opossum conserved sequences emphasizes the importance of marsupial genomes for phylogenetic footprinting-based motif discovery strategies. The BRPR measure is expected to help select genome combinations for optimal performance of these algorithms. Finally, although the etiology of the microRNA upstream increased conservation remains unknown, it is expected to have strong implications for our understanding of regulation of their expression.
Figures



Similar articles
-
ECRbase: database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes.Bioinformatics. 2007 Jan 1;23(1):122-4. doi: 10.1093/bioinformatics/btl546. Epub 2006 Nov 7. Bioinformatics. 2007. PMID: 17090579
-
Homeologs of Brassica SOC1, a central regulator of flowering time, are differentially regulated due to partitioning of evolutionarily conserved transcription factor binding sites in promoters.Mol Phylogenet Evol. 2020 Jun;147:106777. doi: 10.1016/j.ympev.2020.106777. Epub 2020 Feb 29. Mol Phylogenet Evol. 2020. PMID: 32126279
-
Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences.Nature. 2007 May 10;447(7141):167-77. doi: 10.1038/nature05805. Nature. 2007. PMID: 17495919
-
The identification and functional characterisation of conserved regulatory elements in developmental genes.Brief Funct Genomic Proteomic. 2005 Feb;3(4):332-50. doi: 10.1093/bfgp/3.4.332. Brief Funct Genomic Proteomic. 2005. PMID: 15814024 Review.
-
The opossum genome: insights and opportunities from an alternative mammal.Genome Res. 2008 Aug;18(8):1199-215. doi: 10.1101/gr.065326.107. Genome Res. 2008. PMID: 18676819 Free PMC article. Review.
Cited by
-
Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data.PLoS One. 2009;4(4):e5279. doi: 10.1371/journal.pone.0005279. Epub 2009 Apr 23. PLoS One. 2009. PMID: 19390574 Free PMC article.
-
Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors.Bioinformatics. 2008 Sep 1;24(17):1850-7. doi: 10.1093/bioinformatics/btn331. Epub 2008 Jun 27. Bioinformatics. 2008. PMID: 18586699 Free PMC article.
-
Human Lineage-Specific Transcriptional Regulation through GA-Binding Protein Transcription Factor Alpha (GABPa).Mol Biol Evol. 2016 May;33(5):1231-44. doi: 10.1093/molbev/msw007. Epub 2016 Jan 26. Mol Biol Evol. 2016. PMID: 26814189 Free PMC article.
-
Molecular characteristics and clinical implications of serine/arginine-rich splicing factors in human cancer.Aging (Albany NY). 2023 Nov 24;15(22):13287-13311. doi: 10.18632/aging.205241. Epub 2023 Nov 24. Aging (Albany NY). 2023. PMID: 38015716 Free PMC article.
-
Regulation of microRNA expression and function by nuclear receptor signaling.Cell Biosci. 2011 Sep 21;1(1):31. doi: 10.1186/2045-3701-1-31. Cell Biosci. 2011. PMID: 21936947 Free PMC article.
References
-
- Belov K, Deakin JE, Papenfuss AT, Baker ML, Melman SD, Siddle HV, Gouin N, Goode DL, Sargeant TJ, Robinson MD, et al. Reconstructing an ancestral mammalian immune supercomplex from a marsupial major histocompatibility complex. PLoS Biol. 2006;4:e46. doi: 10.1371/journal.pbio.0040046. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

