Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones)
- PMID: 17506894
- PMCID: PMC1904202
- DOI: 10.1186/1471-2164-8-119
Transcriptome analysis of the venom gland of the Mexican scorpion Hadrurus gertschi (Arachnida: Scorpiones)
Abstract
Background: Scorpions like other venomous animals possess a highly specialized organ that produces, secretes and disposes the venom components. In these animals, the last postabdominal segment, named telson, contains a pair of venomous glands connected to the stinger. The isolation of numerous scorpion toxins, along with cDNA-based gene cloning and, more recently, proteomic analyses have provided us with a large collection of venom components sequences. However, all of them are secreted, or at least are predicted to be secretable gene products. Therefore very little is known about the cellular processes that normally take place inside the glands for production of the venom mixture. To gain insights into the scorpion venom gland biology, we have decided to perform a transcriptomic analysis by constructing a cDNA library and conducting a random sequencing screening of the transcripts.
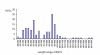
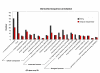
Results: From the cDNA library prepared from a single venom gland of the scorpion Hadrurus gertschi, 160 expressed sequence tags (ESTs) were analyzed. These transcripts were further clustered into 68 unique sequences (20 contigs and 48 singlets), with an average length of 919 bp. Half of the ESTs can be confidentially assigned as homologues of annotated gene products. Annotation of these ESTs, with the aid of Gene Ontology terms and homology to eukaryotic orthologous groups, reveals some cellular processes important for venom gland function; including high protein synthesis, tuned posttranslational processing and trafficking. Nonetheless, the main group of the identified gene products includes ESTs similar to known scorpion toxins or other previously characterized scorpion venom components, which account for nearly 60% of the identified proteins.
Conclusion: To the best of our knowledge this report contains the first transcriptome analysis of genes transcribed by the venomous gland of a scorpion. The data were obtained for the species Hadrurus gertschi, belonging to the family Caraboctonidae. One hundred and sixty ESTs were analyzed, showing enrichment in genes that encode for products similar to known venom components, but also provides the first sketch of cellular components, molecular functions, biological processes and some unique sequences of the scorpion venom gland.
Figures
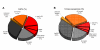



References
-
- Possani LD, Rodríguez de la Vega RC. Scorpion venom peptides. In: Kastin AJ, editor. Handbook of Biologically Active Peptides. San Diego, Academic Press; 2006. pp. 339–354.
-
- Ramanaiah M, Parthasarathy PR, Venkaiah B. Purification and properties of phospholipase A2 from the venom of scorpion, (Heterometrus fulvipes) Biochem Int. 1990;20:931–940. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

