Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus
- PMID: 17507487
- PMCID: PMC1951306
- DOI: 10.1128/JVI.00255-07
Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus
Abstract
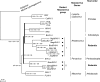
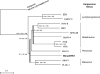
Rodent herpesviruses such as murine cytomegalovirus (host, Mus musculus), rat cytomegalovirus (host, Rattus norvegicus), and murine gammaherpesvirus 68 (hosts, Apodemus species) are important tools for the experimental study of human herpesvirus diseases. However, alphaherpesviruses, roseoloviruses, and lymphocryptoviruses, as well as rhadinoviruses, that naturally infect Mus musculus (house mouse) and other Old World mice are unknown. To identify hitherto-unknown rodent-associated herpesviruses, we captured M. musculus, R. norvegicus, and 14 other rodent species in several locations in Germany, the United Kingdom, and Thailand. Samples of trigeminal ganglia, dorsal root ganglia, brains, spleens, and other organs, as well as blood, were analyzed with a degenerate panherpesvirus PCR targeting the DNA polymerase (DPOL) gene. Herpesvirus-positive samples were subjected to a second degenerate PCR targeting the glycoprotein B (gB) gene. The sequences located between the partial DPOL and gB sequences were amplified by long-distance PCR and sequenced, resulting in a contiguous sequence of approximately 3.5 kbp. By DPOL PCR, we detected 17 novel betaherpesviruses and 21 novel gammaherpesviruses but no alphaherpesvirus. Of these 38 novel herpesviruses, 14 were successfully analyzed by the complete bigenic approach. Most importantly, the first gammaherpesvirus of Mus musculus was discovered (Mus musculus rhadinovirus 1 [MmusRHV1]). This virus is a member of a novel group of rodent gammaherpesviruses, which is clearly distinct from murine herpesvirus 68-like rodent gammaherpesviruses. Multigenic phylogenetic analysis, using an 8-kbp locus, revealed that MmusRHV1 diverged from the other gammaherpesviruses soon after the evolutionary separation of Epstein-Barr virus-like lymphocryptoviruses from human herpesvirus 8-like rhadinoviruses and alcelaphine herpesvirus 1-like macaviruses.
Figures


References
-
- Becker, S. D., M. Bennett, J. P. Stewart, and J. L. Hurst. 2007. Serological survey of virus infection among wild house mice (Mus domesticus) in the UK. Lab. Anim. 41:229-238. - PubMed
-
- Beisser, P. S., S. J. F. Kaptein, E. Beuken, C. A. Bruggeman, and C. Vink. 1998. The Maastricht strain and England strain of rat cytomegalovirus represent different betaherpesvirus species rather than strains. Virology 246:341-351. - PubMed
-
- Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J. Gen. Virol. 84:111-113. - PubMed
-
- Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free-living small rodents. Acta. Virol. 24:468. - PubMed
-
- Booth, T. W., A. A. Scalzo, C. Carrello, P. A. Lyons, H. E. Farrell, G. R. Singleton, and G. R. Shellam. 1993. Molecular and biological characterization of new strains of murine cytomegalovirus isolated from wild mice. Arch. Virol. 132:209-220. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

