Zic1 and Zic3 regulate medial forebrain development through expansion of neuronal progenitors
- PMID: 17507568
- PMCID: PMC6672357
- DOI: 10.1523/JNEUROSCI.4046-06.2007
Zic1 and Zic3 regulate medial forebrain development through expansion of neuronal progenitors
Abstract
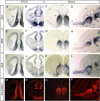
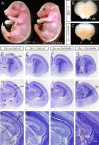
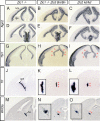
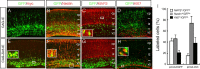
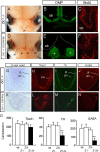
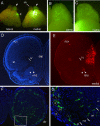
The medial telencephalon is a source of neurons that follow distinct tangential trajectories of migration to various structures such as the cerebral cortex, striatum, and olfactory bulb. In the present study, we characterized the forebrain anomalies in Zic1/Zic3 compound mutant mice. Zic1 and Zic3 were strongly expressed in the medial structures, including the septum, medial cerebral cortex, and choroid plexus. Mice homozygous for the Zic1 mutant allele together with the null Zic3 allele showed medial forebrain defects, which were not obvious in either Zic1 or Zic3 single mutants. Absence of both Zic1 and Zic3 caused hypoplasia of the hippocampus, septum, and olfactory bulb. Analysis of the cell cycle revealed that the cell cycle exit rate was increased in the septa of double mutants. Misexpression of Zic3 in the ventricular layer of the cerebral cortex inhibited neuronal differentiation. These results indicated that both Zic1 and Zic3 function in maintaining neural precursor cells in an undifferentiated state. The functions of these genes may be essential to increasing neural cell numbers regionally in the medial telencephalon and to proper mediolateral patterning of the telencephalon.
Figures



Similar articles
-
Zic2 and Zic3 synergistically control neurulation and segmentation of paraxial mesoderm in mouse embryo.Dev Biol. 2007 Jun 15;306(2):669-84. doi: 10.1016/j.ydbio.2007.04.003. Epub 2007 Apr 12. Dev Biol. 2007. PMID: 17490632
-
Pax6 regulates regional development and neuronal migration in the cerebral cortex.Dev Biol. 2003 Mar 1;255(1):151-63. doi: 10.1016/s0012-1606(02)00046-5. Dev Biol. 2003. PMID: 12618140
-
Zic deficiency in the cortical marginal zone and meninges results in cortical lamination defects resembling those in type II lissencephaly.J Neurosci. 2008 Apr 30;28(18):4712-25. doi: 10.1523/JNEUROSCI.5735-07.2008. J Neurosci. 2008. PMID: 18448648 Free PMC article.
-
Transcriptional regulation of tangential neuronal migration in the developing forebrain.Curr Opin Neurobiol. 2009 Apr;19(2):139-45. doi: 10.1016/j.conb.2009.04.005. Epub 2009 May 8. Curr Opin Neurobiol. 2009. PMID: 19428236 Review.
-
The role of ZIC3 in vertebrate development.Cytogenet Genome Res. 2002;99(1-4):229-35. doi: 10.1159/000071598. Cytogenet Genome Res. 2002. PMID: 12900569 Review.
Cited by
-
Zic2 hypomorphic mutant mice as a schizophrenia model and ZIC2 mutations identified in schizophrenia patients.Sci Rep. 2011;1:16. doi: 10.1038/srep00016. Epub 2011 Jun 17. Sci Rep. 2011. PMID: 22355535 Free PMC article.
-
A Single-Cell Transcriptomic Analysis of the Mouse Hippocampus After Voluntary Exercise.Mol Neurobiol. 2024 Aug;61(8):5628-5645. doi: 10.1007/s12035-023-03869-9. Epub 2024 Jan 13. Mol Neurobiol. 2024. PMID: 38217668 Free PMC article.
-
Zic-Proteins Are Repressors of Dopaminergic Forebrain Fate in Mice and C. elegans.J Neurosci. 2017 Nov 1;37(44):10611-10623. doi: 10.1523/JNEUROSCI.3888-16.2017. Epub 2017 Sep 29. J Neurosci. 2017. PMID: 28972122 Free PMC article.
-
Sequencing-based study of neural induction of human dental pulp stem cells.Hum Cell. 2024 Nov;37(6):1638-1648. doi: 10.1007/s13577-024-01121-7. Epub 2024 Aug 29. Hum Cell. 2024. PMID: 39210197
-
Development of the mammalian main olfactory bulb.Development. 2022 Feb 1;149(3):dev200210. doi: 10.1242/dev.200210. Epub 2022 Feb 11. Development. 2022. PMID: 35147186 Free PMC article. Review.
References
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. - PubMed
-
- Anderson SA, Qiu M, Bulfore A, Eisenstat DD, Meneses, Pederson S, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b;19:27–37. - PubMed
-
- Anderson SA, Marin O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. - PubMed
-
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
