Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae
- PMID: 17507646
- PMCID: PMC1949386
- DOI: 10.1091/mbc.e07-03-0274
Probing the membrane environment of the TOR kinases reveals functional interactions between TORC1, actin, and membrane trafficking in Saccharomyces cerevisiae
Abstract
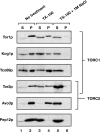
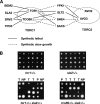
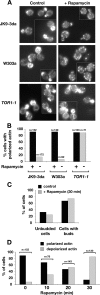
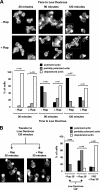
The TOR kinases are regulators of growth in eukaryotic cells that assemble into two distinct protein complexes, TORC1 and TORC2, where TORC1 is inhibited by the antibiotic rapamycin. Present models favor a view wherein TORC1 regulates cell mass accumulation, and TORC2 regulates spatial aspects of growth, including organization of the actin cytoskeleton. Here, we demonstrate that in yeast both TORC1 and TORC2 fractionate with a novel form of detergent-resistant membranes that are distinct from detergent-resistant plasma membrane "rafts." Proteomic analysis of these TOR-associated membranes revealed the presence of regulators of endocytosis and the actin cytoskeleton. Genetic analyses revealed a significant number of interactions between these components and TORC1, demonstrating a functional link between TORC1 and actin/endocytosis-related genes. Moreover, we found that inhibition of TORC1 by rapamycin 1) disrupted actin polarization, 2) delayed actin repolarization after glucose starvation, and 3) delayed accumulation of lucifer yellow within the vacuole. By combining our genetic results with database mining, we constructed a map of interactions that led to the identification of additional genetic interactions between TORC1 and components involved in membrane trafficking. Together, these results reveal the broad scope of cellular processes influenced by TORC1, and they underscore the functional overlap between TORC1 and TORC2.
Figures





References
-
- Araki T., Uesono Y., Oguchi T., Toh E. A. LAS24/KOG1, a component of the TOR complex 1 (TORC1), is needed for resistance to local anesthetic tetracaine and normal distribution of actin cytoskeleton in yeast. Genes Genet. Syst. 2005;80:325–343. - PubMed
-
- Berven L. A., Willard F. S., Crouch M. F. Role of the p70(S6K) pathway in regulating the actin cytoskeleton and cell migration. Exp. Cell Res. 2004;296:183–195. - PubMed
-
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

