Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3'-UTR
- PMID: 17507659
- PMCID: PMC1894925
- DOI: 10.1261/rna.577707
Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3'-UTR
Abstract
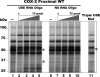
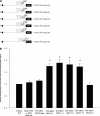
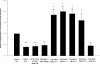
Two cyclooxygenase (COX) enzymes, COX-1 and COX-2, are present in human cells. While COX-1 is constitutively expressed, COX-2 is inducible and up-regulated in response to many signals. Since increased transcriptional activity accounts for only part of COX-2 up-regulation, we chose to explore other RNA processing mechanisms in the regulation of this gene. Previously, we showed that COX-2 is regulated by alternative polyadenylation, and that the COX-2 proximal polyadenylation signal contains auxiliary upstream sequence elements (USEs) that are very important in efficient polyadenylation. To explore trans-acting protein factors interacting with these cis-acting RNA elements, we performed pull-down assays with HeLa nuclear extract and biotinylated RNA oligonucleotides representing COX-2 USEs. We identified PSF, p54(nrb), PTB, and U1A as proteins specifically bound to the COX-2 USEs. We further explored their participation in polyadenylation using MS2 phage coat protein-MS2 RNA binding site tethering assays, and found that tethering any of these four proteins to the COX-2 USE mutant RNA can compensate for these cis-acting elements. Finally, we suggest that these proteins (p54(nrb), PTB, PSF, and U1A) may interact as a complex since immunoprecipitations of the transfected MS2 fusion proteins coprecipitate the other proteins.
Figures


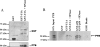


Similar articles
-
A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein.J Virol. 2005 Apr;79(7):4270-88. doi: 10.1128/JVI.79.7.4270-4288.2005. J Virol. 2005. PMID: 15767428 Free PMC article.
-
p54nrb is a component of the snRNP-free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation.RNA. 2006 Jan;12(1):111-21. doi: 10.1261/rna.2213506. RNA. 2006. PMID: 16373496 Free PMC article.
-
The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha.J Biol Chem. 2008 Jan 4;283(1):57-65. doi: 10.1074/jbc.M705286200. Epub 2007 Oct 26. J Biol Chem. 2008. PMID: 17965020
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
-
Complexity of COX-2 gene regulation.Biochem Soc Trans. 2008 Jun;36(Pt 3):543-5. doi: 10.1042/BST0360543. Biochem Soc Trans. 2008. PMID: 18482003 Review.
Cited by
-
ADAR editing in double-stranded UTRs and other noncoding RNA sequences.Trends Biochem Sci. 2010 Jul;35(7):377-83. doi: 10.1016/j.tibs.2010.02.008. Epub 2010 Apr 8. Trends Biochem Sci. 2010. PMID: 20382028 Free PMC article. Review.
-
αCP Poly(C) binding proteins act as global regulators of alternative polyadenylation.Mol Cell Biol. 2013 Jul;33(13):2560-73. doi: 10.1128/MCB.01380-12. Epub 2013 Apr 29. Mol Cell Biol. 2013. PMID: 23629627 Free PMC article.
-
An RNA-protein complex links enhanced nuclear 3' processing with cytoplasmic mRNA stabilization.EMBO J. 2011 May 27;30(13):2622-33. doi: 10.1038/emboj.2011.171. EMBO J. 2011. PMID: 21623344 Free PMC article.
-
Nucleophosmin deposition during mRNA 3' end processing influences poly(A) tail length.EMBO J. 2011 Aug 5;30(19):3994-4005. doi: 10.1038/emboj.2011.272. EMBO J. 2011. PMID: 21822216 Free PMC article.
-
Consensus PP1 binding motifs regulate transcriptional corepression and alternative RNA splicing activities of the steroid receptor coregulators, p54nrb and PSF.Mol Endocrinol. 2011 Jul;25(7):1197-210. doi: 10.1210/me.2010-0517. Epub 2011 May 12. Mol Endocrinol. 2011. PMID: 21566083 Free PMC article.
References
-
- Bazan, N.G. COX-2 as a multifunctional neuronal modulator. Nat. Med. 2001;7:414–415. - PubMed
-
- Bentley, D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. - PubMed
-
- Bishop-Bailey, D., Calatayud, S., Warner, T.D., Hla, T., Mitchell, J.A. Prostaglandins and the regulation of tumor growth. J. Environ. Pathol. Toxicol. Oncol. 2002;21:93–101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
