The in silico map-based cloning of Pi36, a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus
- PMID: 17507669
- PMCID: PMC1950653
- DOI: 10.1534/genetics.107.075465
The in silico map-based cloning of Pi36, a rice coiled-coil nucleotide-binding site leucine-rich repeat gene that confers race-specific resistance to the blast fungus
Abstract
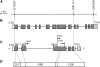
The indica rice variety Kasalath carries Pi36, a gene that determines resistance to Chinese isolates of rice blast and that has been located to a 17-kb interval on chromosome 8. The genomic sequence of the reference japonica variety Nipponbare was used for an in silico prediction of the resistance (R) gene content of the interval and hence for the identification of candidate gene(s) for Pi36. Three such sequences, which all had both a nucleotide-binding site and a leucine-rich repeat motif, were present. The three candidate genes were amplified from the genomic DNA of a number of varieties by long-range PCR, and the resulting amplicons were inserted into pCAMBIA1300 and/or pYLTAC27 vectors to determine sequence polymorphisms correlated to the resistance phenotype and to perform transgenic complementation tests. Constructs containing each candidate gene were transformed into the blast-susceptible variety Q1063, which allowed the identification of Pi36-3 as the functional gene, with the other two candidates being probable pseudogenes. The Pi36-encoded protein is composed of 1056 amino acids, with a single substitution event (Asp to Ser) at residue 590 associated with the resistant phenotype. Pi36 is a single-copy gene in rice and is more closely related to the barley powdery mildew resistance genes Mla1 and Mla6 than to the rice blast R genes Pita, Pib, Pi9, and Piz-t. An RT-PCR analysis showed that Pi36 is constitutively expressed in Kasalath.
Figures






References
-
- Bent, A. F., B. N. Kunkel, D. Dahlbeck, K. L. Brown, R. Schmidt et al., 1994. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

