Defective decapentaplegic signaling results in heart overgrowth and reduced cardiac output in Drosophila
- PMID: 17507674
- PMCID: PMC1931542
- DOI: 10.1534/genetics.107.073569
Defective decapentaplegic signaling results in heart overgrowth and reduced cardiac output in Drosophila
Abstract
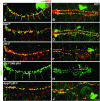
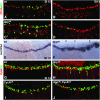
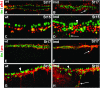
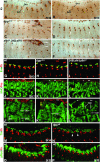
During germ-band extension, Decapentaplegic (Dpp) signals from the dorsal ectoderm to maintain Tinman (Tin) expression in the underlying mesoderm. This signal specifies the cardiac field, and homologous genes (BMP2/4 and Nkx2.5) perform this function in mammals. We showed previously that a second Dpp signal from the dorsal ectoderm restricts the number of pericardial cells expressing the transcription factor Zfh1. Here we report that, via Zfh1, the second Dpp signal restricts the number of Odd-skipped-expressing and the number of Tin-expressing pericardial cells. Dpp also represses Tin expression independently of Zfh1, implicating a feed-forward mechanism in the regulation of Tin pericardial cell number. In the adjacent dorsal muscles, Dpp has the opposite effect. Dpp maintains Krüppel and Even-skipped expression required for muscle development. Our data show that Dpp refines the cardiac field by limiting the number of pericardial cells. This maintains the boundary between pericardial and dorsal muscle cells and defines the size of the heart. In the absence of the second Dpp signal, pericardial cells overgrow and this significantly reduces larval cardiac output. Our study suggests the existence of a second round of BMP signaling in mammalian heart development and that perhaps defects in this signal play a role in congenital heart defects.
Figures





Similar articles
-
Embryonic enhancers in the dpp disk region regulate a second round of Dpp signaling from the dorsal ectoderm to the mesoderm that represses Zfh-1 expression in a subset of pericardial cells.Dev Biol. 2003 Oct 1;262(1):137-51. doi: 10.1016/s0012-1606(03)00350-6. Dev Biol. 2003. PMID: 14512024
-
Antagonistic function of Lmd and Zfh1 fine tunes cell fate decisions in the Twi and Tin positive mesoderm of Drosophila melanogaster.Dev Biol. 2009 Feb 15;326(2):444-55. doi: 10.1016/j.ydbio.2008.10.041. Epub 2008 Nov 11. Dev Biol. 2009. PMID: 19028484
-
Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors.Dev Biol. 2001 Oct 1;238(1):13-26. doi: 10.1006/dbio.2001.0397. Dev Biol. 2001. PMID: 11783990
-
TGF-beta family signal transduction in Drosophila development: from Mad to Smads.Dev Biol. 1999 Jun 15;210(2):251-68. doi: 10.1006/dbio.1999.9282. Dev Biol. 1999. PMID: 10357889 Review.
-
Methods to assess Drosophila heart development, function and aging.Methods. 2014 Jun 15;68(1):265-72. doi: 10.1016/j.ymeth.2014.03.031. Epub 2014 Apr 12. Methods. 2014. PMID: 24727147 Free PMC article. Review.
Cited by
-
Salmonella pathogenesis reveals that BMP signaling regulates blood cell homeostasis and immune responses in Drosophila.Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):14952-7. doi: 10.1073/pnas.0808208105. Epub 2008 Sep 24. Proc Natl Acad Sci U S A. 2008. PMID: 18815369 Free PMC article.
-
On the Morphology of the Drosophila Heart.J Cardiovasc Dev Dis. 2016 Apr 12;3(2):15. doi: 10.3390/jcdd3020015. J Cardiovasc Dev Dis. 2016. PMID: 29367564 Free PMC article. Review.
-
A combinatorial enhancer recognized by Mad, TCF and Brinker first activates then represses dpp expression in the posterior spiracles of Drosophila.Dev Biol. 2008 Jan 15;313(2):829-43. doi: 10.1016/j.ydbio.2007.10.021. Epub 2007 Oct 24. Dev Biol. 2008. PMID: 18068697 Free PMC article.
-
Excessive Dpp signaling induces cardial apoptosis through dTAK1 and dJNK during late embryogenesis of Drosophila.J Biomed Sci. 2011 Nov 24;18(1):85. doi: 10.1186/1423-0127-18-85. J Biomed Sci. 2011. PMID: 22114909 Free PMC article.
-
3D chromatin interactions organize Yan chromatin occupancy and repression at the even-skipped locus.Genes Dev. 2013 Nov 1;27(21):2293-8. doi: 10.1101/gad.225789.113. Genes Dev. 2013. PMID: 24186975 Free PMC article.
References
-
- Alvarez, A., W. Shi, B. Wilson and J. Skeath, 2003. pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development 130: 3015–3026. - PubMed
-
- Bour, B., M. O'Brien, W. Lockwood, E. Goldstein, R. Bodmer et al., 1995. dMef2, a transcription factor that is essential for myogenesis. Genes Dev. 9: 730–741. - PubMed
-
- Brand, A., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

