A validated whole-genome association study of efficient food conversion in cattle
- PMID: 17507676
- PMCID: PMC1931545
- DOI: 10.1534/genetics.107.072637
A validated whole-genome association study of efficient food conversion in cattle
Abstract
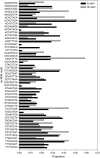
The genetic factors that contribute to efficient food conversion are largely unknown. Several physiological systems are likely to be important, including basal metabolic rate, the generation of ATP, the regulation of growth and development, and the homeostatic control of body mass. Using whole-genome association, we found that DNA variants in or near proteins contributing to the background use of energy of the cell were 10 times as common as those affecting appetite and body-mass homeostasis. In addition, there was a genic contribution from the extracellular matrix and tissue structure, suggesting a trade-off between efficiency and tissue construction. Nevertheless, the largest group consisted of those involved in gene regulation or control of the phenotype. We found that the distribution of micro-RNA motifs was significantly different for the genetic variants associated with residual feed intake than for the genetic variants in total, although the distribution of promoter sequence motifs was not different. This suggests that certain subsets of micro-RNA are more important for the regulation of this trait. Successful validation depended on the sign of the allelic association in different populations rather than on the strength of the initial association or its size of effect.
Figures

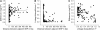
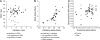

References
-
- Barendse, W. J., 2002. DNA markers for meat tenderness. Patent application WO02064820.
-
- Barendse, W., 2005. The transition from quantitative trait loci to diagnostic test in cattle and other livestock. Aust. J. Exp. Agric. 45: 831–836.
-
- Barendse, W., R. J. Bunch, B. E. Harrison and M. B. Thomas, 2006. The growth hormone 1 GH1:c.457C>G mutation is associated with relative fat distribution in intra-muscular and rump fat in a large sample of Australian feedlot cattle. Anim. Genet. 37: 211–214. - PubMed
-
- Bejerano, G., M. Pheasant, I. Makunin, S. Stephen, W. J. Kent et al., 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

